¶ Introduction
You can easily deposit a new structure in 3decision via the structure registration workflow in the User Interface (UI). Both EXPERIMENTAL and PREDICTED structures can be deposited.
Structure registration via the 3decision REST API is described here.
¶ Requirements
- Structure file: a PDB-file is mandatory for the structure registration. The file format needs to be compliant with RCSB standards.
- Ligand description: the bond order of proprietary ligands needs to be described by a SMILES string. A ligand depiction is displayed in the workflow to allow for visual validation. The ligand description of public small molecules is taken from the RSCB PDB Chemical Component Dictionary.
- Protein chain identification: each protein chain needs to be identified with a corresponding UniProt id. This information can be defined directly in the structure file (DBREF) or given in the workflow.
Check if your PDB file is RCSB compliant using the REST API endpoint
POST /structure-files/validationdescribed here.
¶ Tutorial
This section describes step-by-step how to register a new structure via the 3decision UI.
You will learn how to access the 3decision structure registration page, and complete all steps to correctly register your structure.
We will give an example of the registration of a GPCR structure (PDB code: 5tzy), in complex with two ligands: one publically available (7OS), and one that mimicks the situation you experience with a proprietary ligand (LIG).
To follow along this example, you can download the Structure File here: 5tzy_lig.pdb.
¶ 1. Access Structure Registration Page from UI
- From the 3decision Homepage, click on the "upload arrow" button on the left-side menu:

- A new tab opens with the Structure Registration Page. Select the tile "New Structure".
You are directed to the structure registration page. On top, you have an overview of the full workflow, from the file upload to submission of a structure.
¶ 2. Upload a Structure File
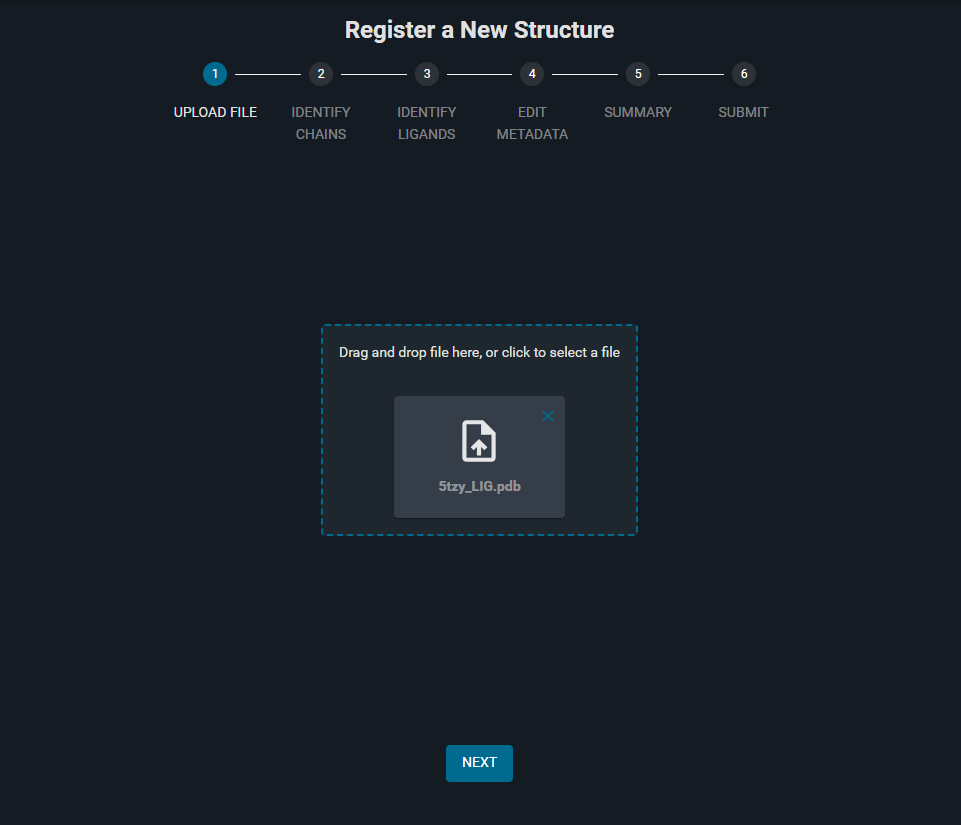
- Drag and drop the Structure File in the dropzone (in the dotted rectangle at the center of the page). Alternatively, click in the dropzone to select the file to upload from folder.
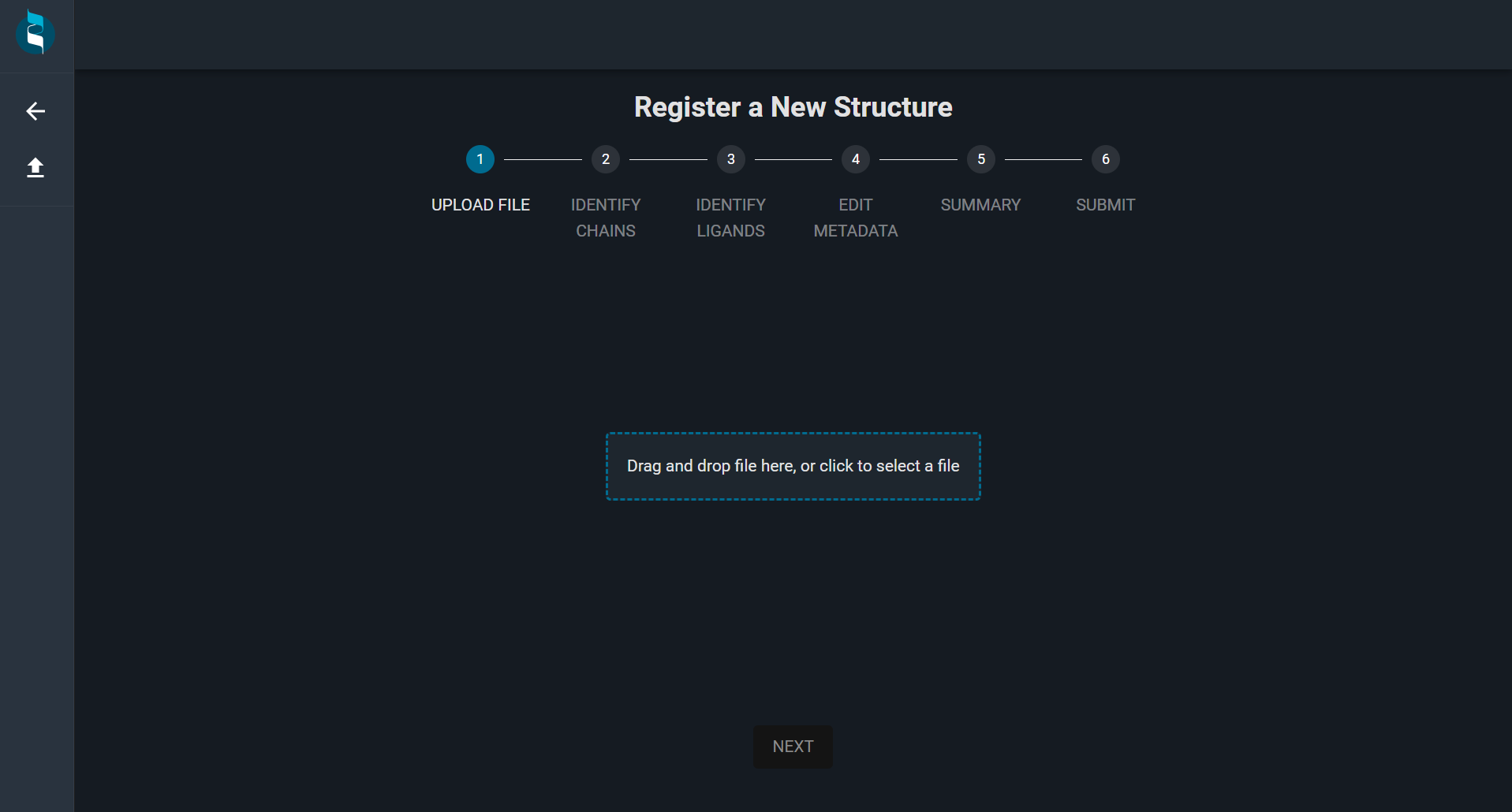
- After the upload and parsing of the file is completed, Click on the blue button "NEXT" at the bottom of the page
¶ 3. Identify Chains
The automatically identified chains with corresponding Biomolecule code and residue ranges are listed on this page. In this tutorial, the data was available in the DBREF section of the PDB file. You can edit these fields if the information is missing.
- The Biomolecule codes (Uniprot identifier) and the residue ranges are as follows:
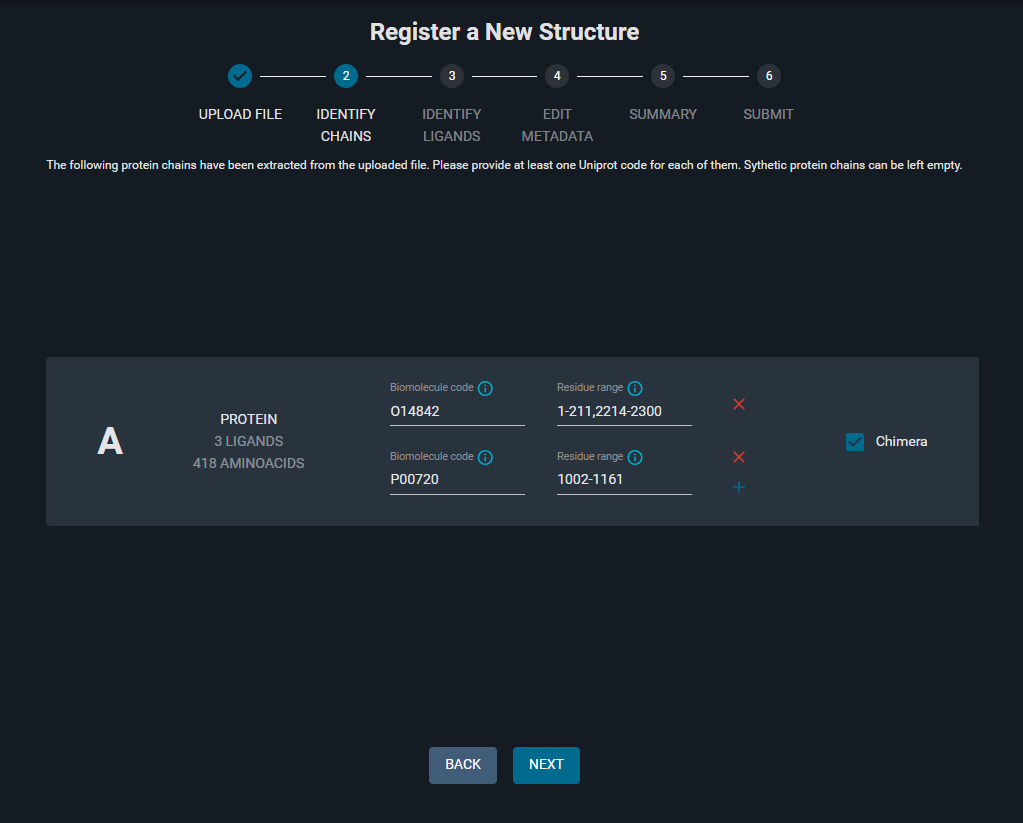
Notice that in this case, you have a Chimera protein structure, as indicated by the option "Chimera" toggled on (on the right), that gives you the possibility to specify different Biomolecule codes for different residue ranges on the same chain.
- Click "NEXT" at the bottom of the page
Chimeric protein chains can be defined by checking the option "Chimera". Multiple residue ranges on the same protein chain can thus be defined by unique UniProt identifiers.
Synthetic protein chains do not have corresponding Uniprot identifiers and the "Biomolecule code" field can thus be left empty. The protein chain sequence will be registered as a new sequence in 3decision and assigned a unique biomolecule code, in the format
PRO_XXXXXX. If you want, you can register a user sequence using the REST API and then use the generated Biomol_code in this step.
¶ 4. Identify Ligands
A list of the identified small molecules (all HETATM), with the corresponding 2D structure representation and SMILES string, is shown on this page. You can edit the SMILES, and specify an INTERNAL ID for each ligand.
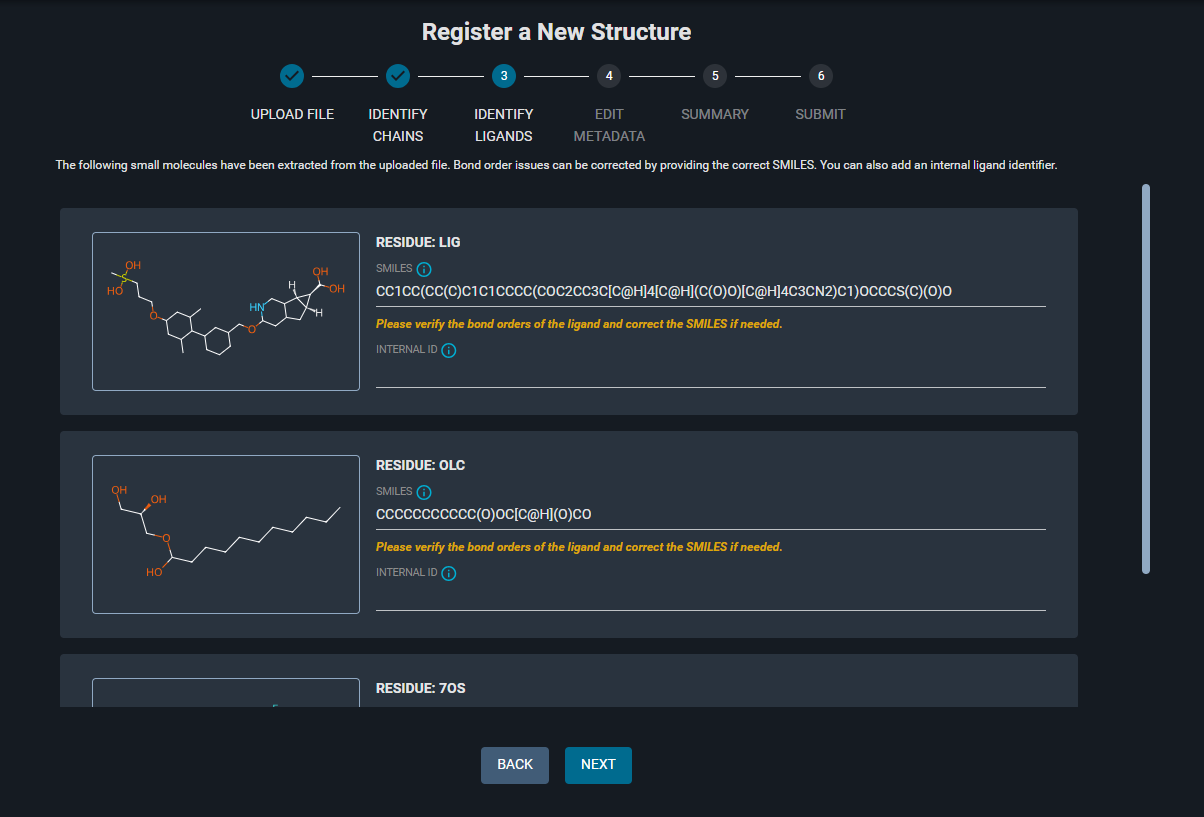
- Check the ligand
LIG: the structure is not correct (double bonds missing).
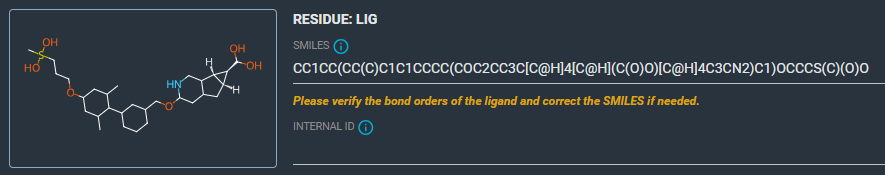
- Correct the SMILES for LIG, using this string:
CC1=CC(OCCCS(C)(=O)=O)=CC(C)=C1C1=CC(COC2=NC=C3[C@H]4[C@@H](CC3=C2)[C@@H]4C(O)=O)=CC=C1. The 2D structure of the ligand is now correct.

- For LIG, add the Internal ID:
LIG-XXX

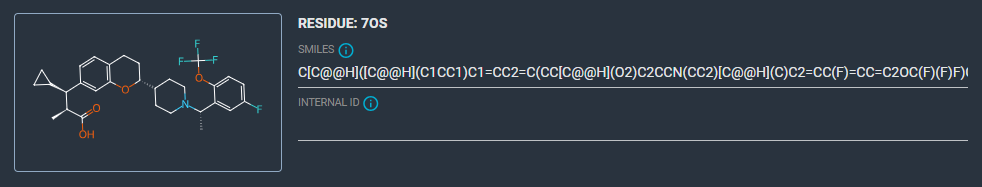
- Check the ligand
7OS: it has the correct structure.
- Click "NEXT" at the bottom of the page
¶ 5. Edit Metadata
On this page you can manage the structure metadata, regulate the access to the structure, and define its relation with other structures in the database.
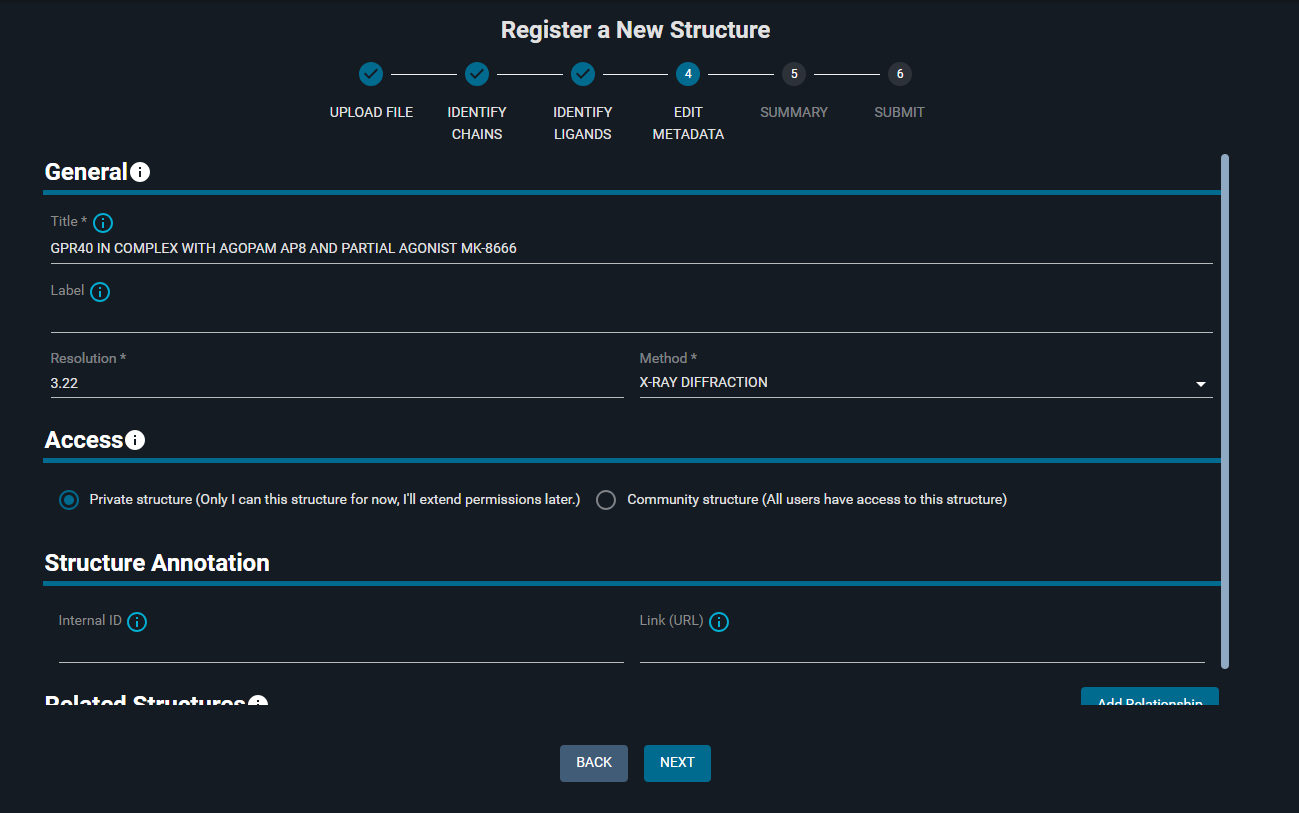
¶ General: edit Structure Metadata
The general information on the structure you find here are parsed from the PDB header of the Structure File.
- Change the title to:
REGISTRATION TEST - (Optional: add a Label to the structure)
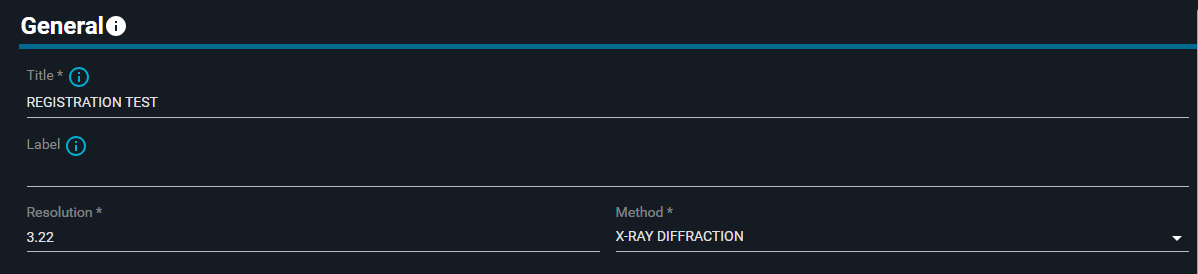
The Methods options available are: X-RAY DIFFRACTION, SOLUTION NMR, ELECTRON MICROSCOPY, Model)
¶ Access: share the structure
You can decide whether to keep the structure private with the option "Private structure" toggled on, or share it with all the users by toggling the "Community structure" option.
- Keep the "Private structure" option toggled on
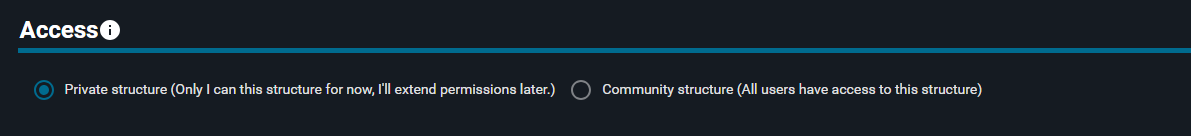
If you keep the structure private, you can regulate the access permissions after registration using 3decision privileges),
¶ Structure Annotation
You can annotate your structure with an Internal ID and a link.
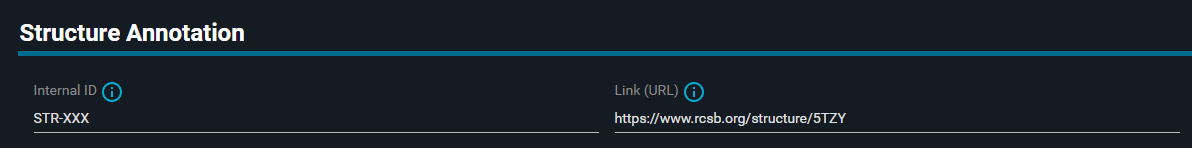
- Add the following "Internal ID":
STR-XXX - (Optional: in the "Link (URL)" paste the link:
https://www.rcsb.org/structure/5TZY.)
TIP: you can use this field to put a link to the ELN for your proprietary structure.
Any other annotation can be added to the structure after registration using the 3decision REST API, as described here.
¶ Related Structures: add relationships
In this section, you can specify if the new structure is related to other structures in the database.
- Click on "Add Relationship" button
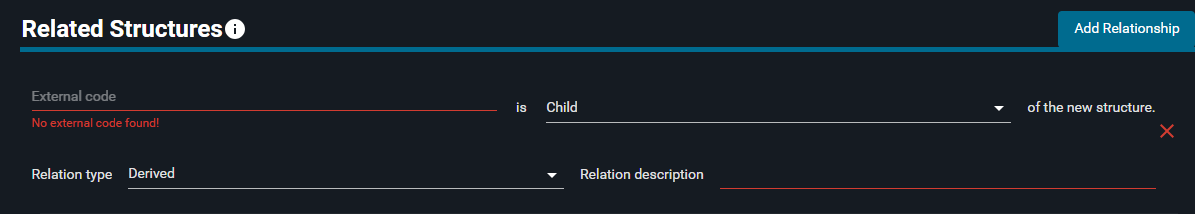
- In the "External code" field, type
5TZY. From the dropdown menu that appears, select "5tzy" - Click on "Child", and from the dropdown menu select "Parent"
- In the "Relation type" section, click on "Derived" and select "Refined" from the list
- In the "Relation description" part, write:
Registration test
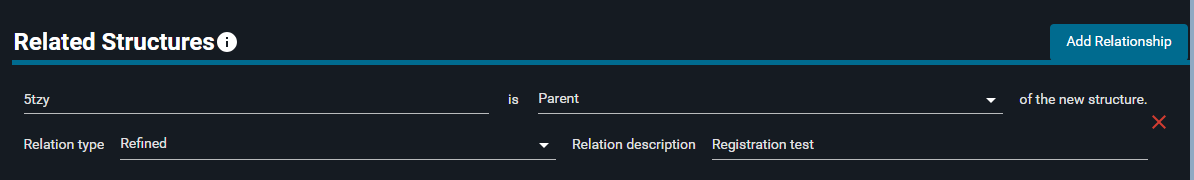
- Click "NEXT" at the bottom of the page
The available hierarchical types are:
ParentandChild.
The available relation types are:Derived,Bioassembly, andRefined.
The structure relationships defined here will be shown in the Information Browser, allowing easy browsing between related structures.
¶ 6. Finalize the Structure Registration
The SUMMARY page give an overview of the information given in the registration process. You can check that the provided information is correct. If needed you can use the "BACK" button to go back and edit any data.
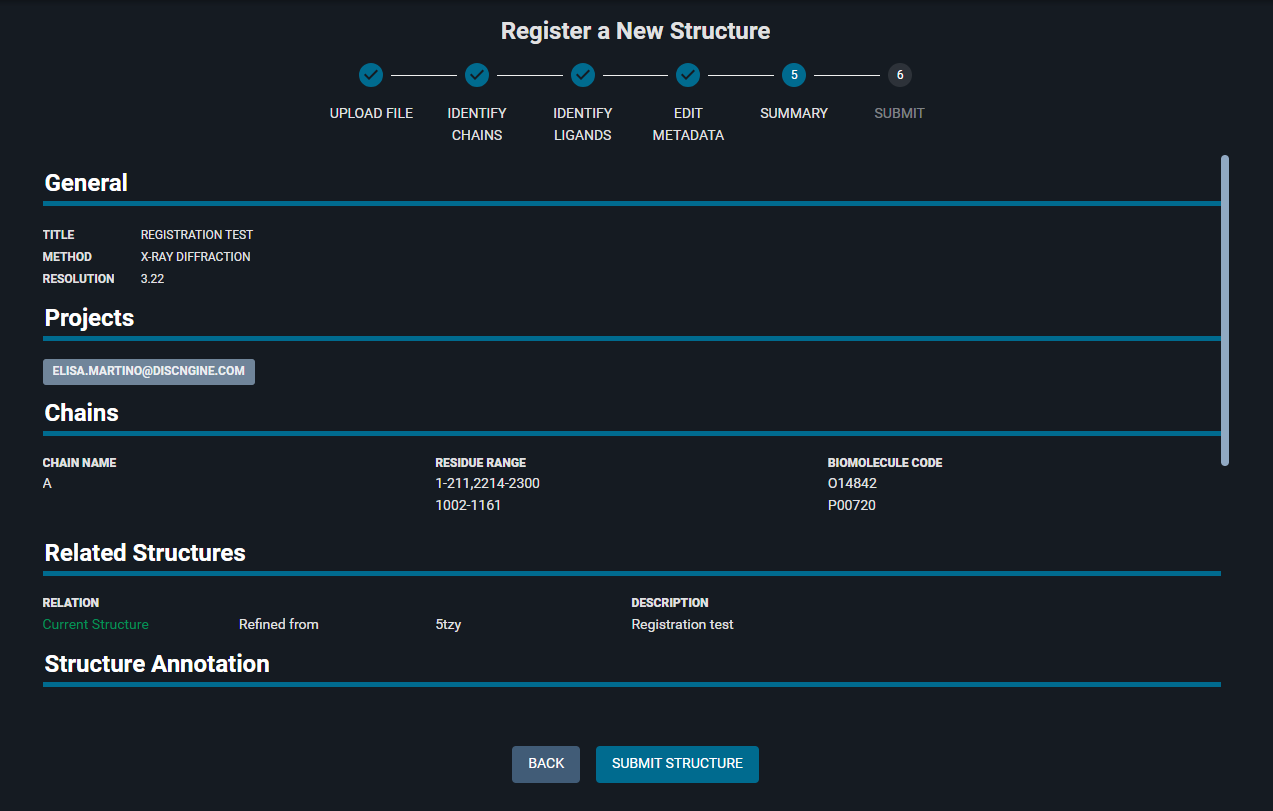
- Click "SUBMIT STRUCTURE" at the bottom of the page.
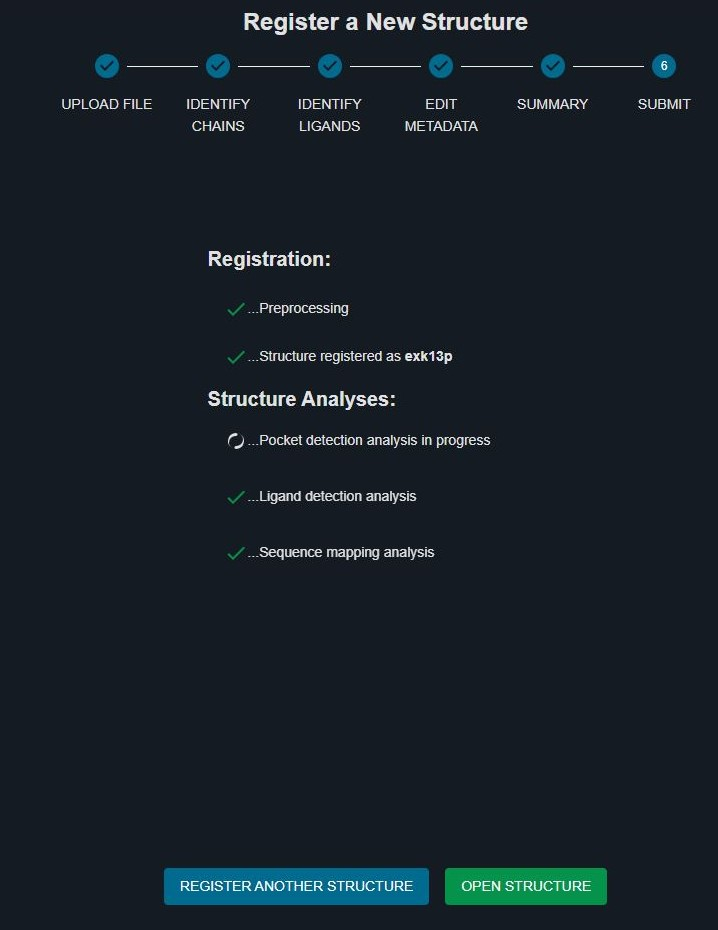
You are now directed to the SUBMIT page and will see the list of the different structure registration and analysis steps and their status:

When the structure has been succesfully registered structure, it's given a unique 6-digits external code (yours will differ from the one reported in this example):
¶ 7. Open the new structure
- Once all structure analyses have been completed, click on "OPEN STRUCTURE" (green button at the bottom of the page)
A new 3decision tab will open and you can see the structure in the 3D Viewer on the right.
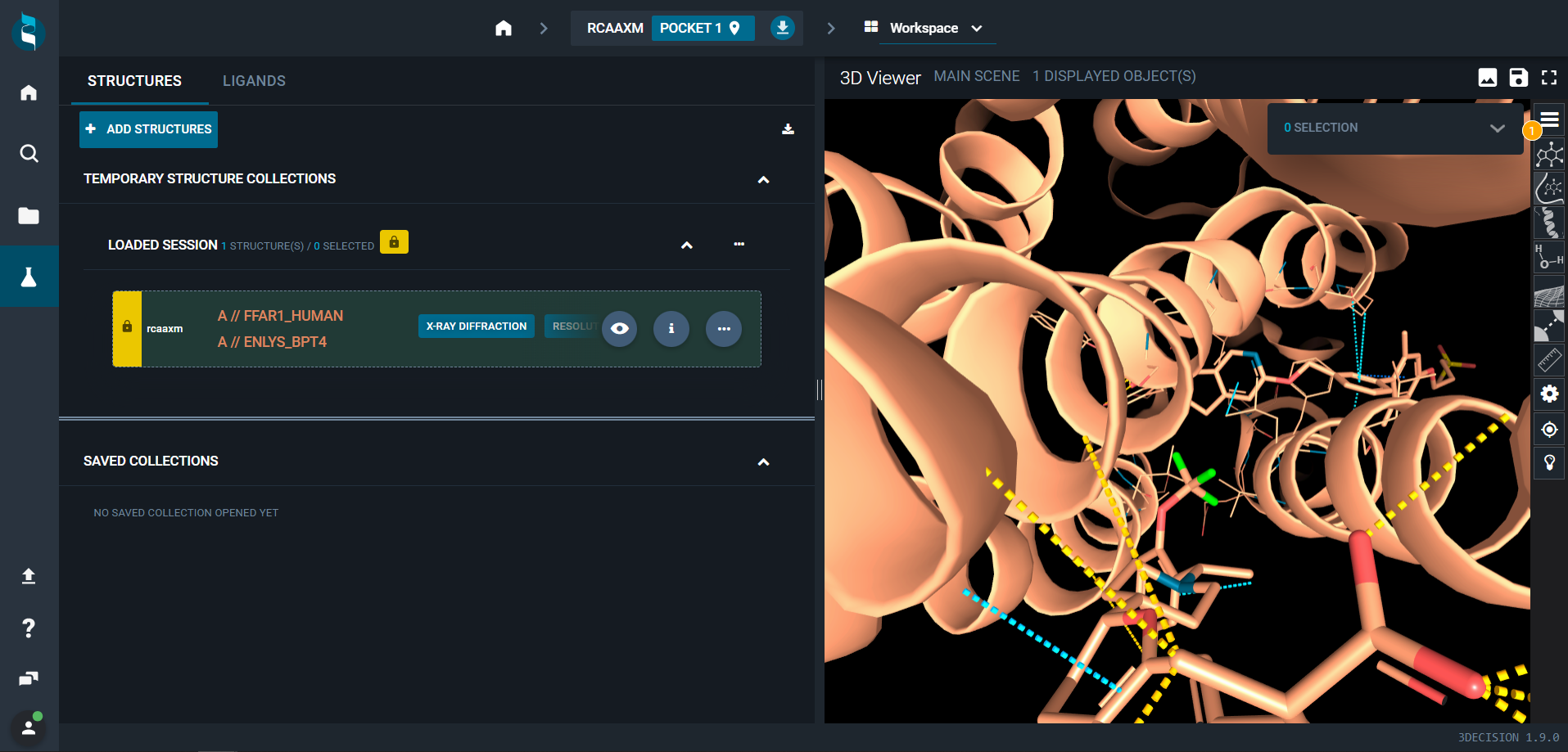
The structure was correctly included in the 3decision database, and is now searchable and ready to work with!