The 3decision API can be used to register proprietary structures (both experimental and in silico models).
Publicly available structures from the RCSB PDB are automatically registred in the database on a weekly basis.
This section describes the requirements for performing the structure import, and how to use 3decision API endpoints to register new structures in the database. Additionally, instructions for checking the status of the registration are reported.
¶ Structure Import Requirements
To import a structure in the 3decision database, you need:
- the structure file to register in one of the following formats:
.pdb,.pdb.gz,.ent,.ent.gz,.cif,.cif.gz. - the registration payload containing structure details and (recommended, not required) structure metadata (more details in the section below).
PDB Structure Files should be RCSB-compliant for a successful registration (reference for compliance here).
You can use thePOST/ structure-files/validationendpoint to check the compliance of your Structure File with the 3decision registration process. Details on how to use this endpoint are reportend in this page of the user guide.
¶ Registration Payload
All the structure details of the registration payload are described in the Schema of the POST /structure-registration endpoint request body:
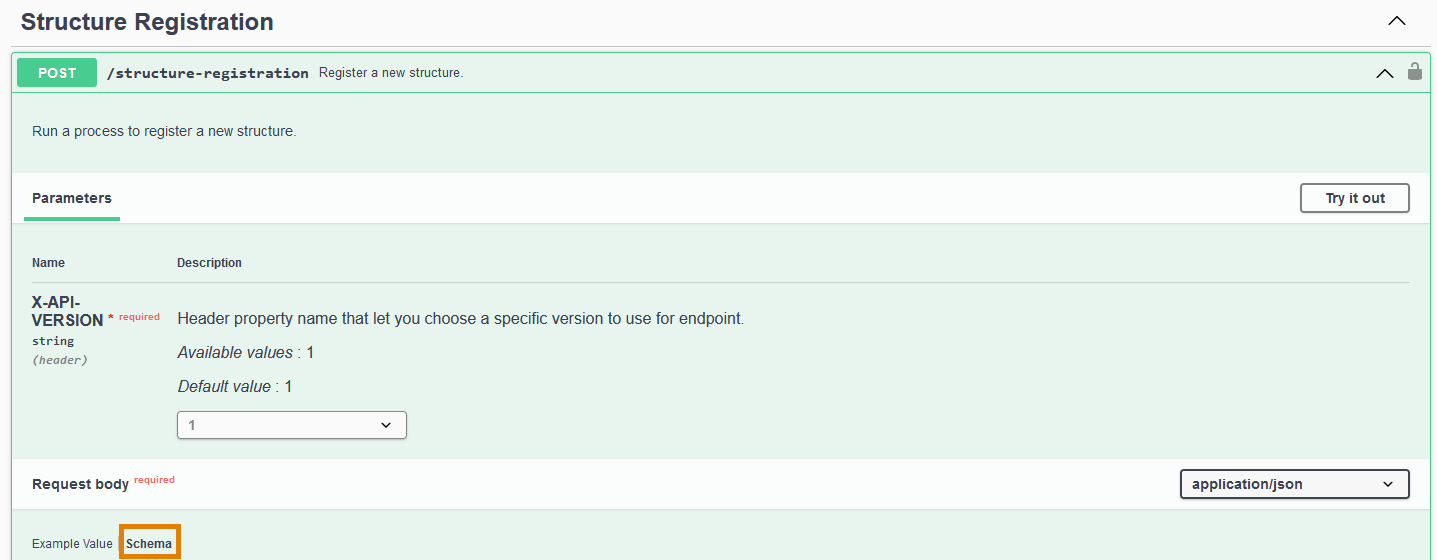
In the table, each field of the payload is detailed:
| Name ( * = required) |
Type | Description |
|---|---|---|
experimentalMethod* |
string |
Experimental method used to determine the atomic structure. It must match existing already experimental methods. They can be found with the GET /experimental-methods endpoint (most frequent are: X-RAY DIFFRACTION, ELECTRON MICROSCOPY, SOLUTION NMR, MODEL) |
commit |
boolean |
if True, the data is saved in database, otherwise, the registration will be a dry run: the registration will be run, but the structure won't be persisted. |
resolution* |
number |
Resolution obtained during structure determination, in case of SOLUTION NMR or MODEL experimental methods, use 0 as resolution. |
filePath* |
string |
File to the structure file on the server, obtained after uploading the structure file (POST /structures/upload endpoint) |
title* |
string |
Structure title which will be displayed in 3decision |
label |
string |
Set a label to the structure (more details below) |
projectIds |
list[number] |
List of 3decision project IDs to which the structure is linked. All project IDs can be found using the GET /projects endpoint. A structure must be added to the “community” project to make it accessible to all users ("ProjectIds": [2]) |
annotation |
{label*:string,link: string,typeLabel*: string} |
Add an annotation to the structure, link creates a clickable link displayed in structure info pannel, in the 3decision user interface. Internal ID is the only available typeLabel at registration. |
metadata* |
See section below | Information about ligands and chains mapping to protein sequence |
structureRelation |
list[{externalCode*: string,relationType*: string,hierarchyType*: string,relationDescription*: string}] |
List of already registered structures to which the registered structure should be linked. The structure relationship will be shown in the user interface in the Information panel, allowing browsing between related structures. relationType must be one of the following: Refined, Derived, Prepared for docking, Bioassembly. hierarchicalType must be one of the following: Parent, Child. |
Example of annotation payload:
"annotation": {
"label": "11223344",
"link": "https://<company>/compound/11223344",
"typeLabel": "Internal ID"
}
Example of a structure relation payload:
{
"externalCode": "kzyxt7",
"relationType": "Derived",
"hierarchyType": "Parent",
"relationDescription": "The registered structure is derived from the parent structure kzywt7"
}
¶ Label
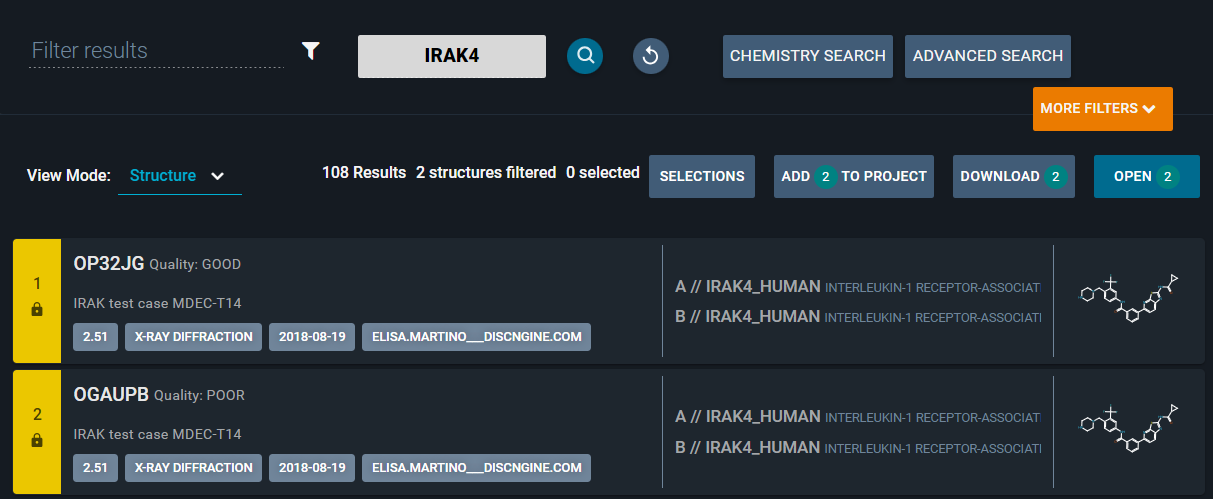
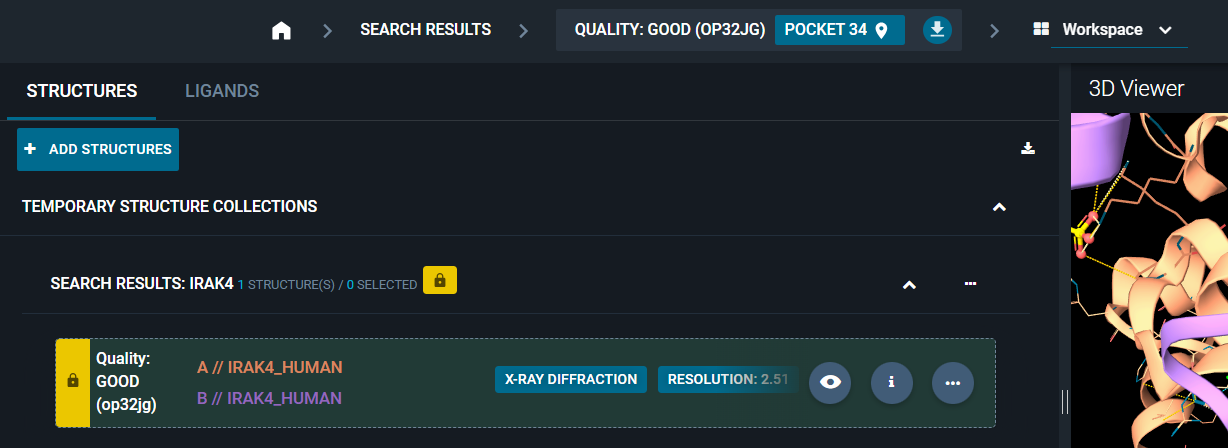
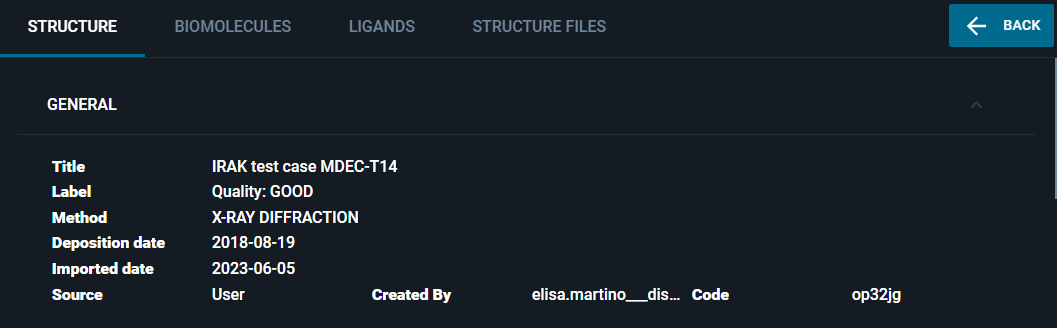
You can add a label to any structure registered in the database, specifying this field in the structure registration payload.
This label will be displayed:
- In the Results Navigation Page (Structure and Ligand View Mode): in the structure/ligand card.
- In the Workspace: in the structure/ligand card.
- In the Information Browser: in the "GENERAL" section.
¶ Structure Metadata
Structure metadatain the payload is composed of 2 (non-required) elements: chains and ligands.
- Chains represent the mapping between structure chains and the protein sequences (Uniprot biomolecules).
- Ligands are used to customize ligands information (with a ligand internal ID), and to correct bond orders in the representation of the molecules.
Types of the metadata ligands and chains are:
"metadata": {
"ligands": list[
{
chain: string,
residueNumber: number,
residueCode: string,
internalId: string,
smiles*: string
}
],
"chains": list[
{
name*: string,
biomolCode*: string,
from: number,
to: number
}
]
}
¶ Example of chain mapping payload
"metadata": {
"chains": [
{"name": ”A”, "biomolCode": ”RBP1_PLAF7”},
{"name": ”B”, "biomolCode": ”RBP1_PLAF7”, "from": 1, "to": 20},
{"name": ”B”, "biomolCode": ”RBP1_DROME”, "from": 23,"to": 56}
]
}
In this example, the two ways to provide chains mapping were used:
- chain A is defined as being RBP1_PLAF7 biomolecule
- chain B is RBP1_PLAF7 from residue 1 to 20 and RBP1_DROME from 23 to 56.
The residues that are not mapped through this input will be covered by a generic mapping: a generic sequence is produced (biomolecule code PRO_XXX by convention)
Automatic chain mapping to biomolecules will be available in an upcoming version.
The threshold for matching a sequence in chain mapping is 80%. With lower similarity score, the sequence deviates too much from the Uniprot one and will result in chain mapping failing.
¶ Example of ligands payload
"metadata": {
"ligands": [
{
"residueCode": "5CV",
"smiles": "CC(C)(CN(C)C)COc1ccc2c(c1)cc([nH]2)C3=CC(=CNC3=O)NC(=O)c4cnn(c4)Cc5ccccc5",
"internalId": "CPD44198296"
}
]
}
During the structure registration, the small molecules are corrected using one of the following approaches:
- When the user provides the residue code (3-letter code of the ligand): the correction is performed using the template SMILES matching the residue code.
- If no matching residue code is found, the SMILES provided by the user are compared and the first matching SMILES is used as template for the correction.
- If no template is provided and no SMILES are matching the processed small molecule, the chemcomp dictionary (references provided by RCSB) is used. This step is only possible for experimental ligands and not for models where the ligand is called LIG.
- If no information is provided and if the ligand is a model called LIG, an automatic bond detection is performed based on the small molecule coordinates.
If a processed ligand can’t be fixed and contains errors, the ligand will not be registered, and the structure will be annotated with this missing ligand.
¶ Registration of a Structure
To register a structure in 3decision from the API, you first need to access and activate the API (instructions in the Access page of this documentation).
Then, a structure can be registered in two steps:
- Upload the structure file (endpoint:
POST /structures/upload) - Submit the structure registration (endpoint:
POST /structure-registration)
For each step, instructions from the 3decision API interface, Curl commands, and Python scripts are reported.
Warning: Structures with less than 100,000 atoms can be registered in the database.
¶ 1. Upload the structure file
The first step to register a structure in the database is to upload the file on the server using the POST /structures/upload endpoint.
This endpoint will return a server file path (required for the second step).
Note that the file name length must be less than or equal to 200 characters.
¶ From the 3decision API interface
- From the activated API interface, go to the
POST /structures/uploadendpoint - Click on "Try it out" to activate the endpoint
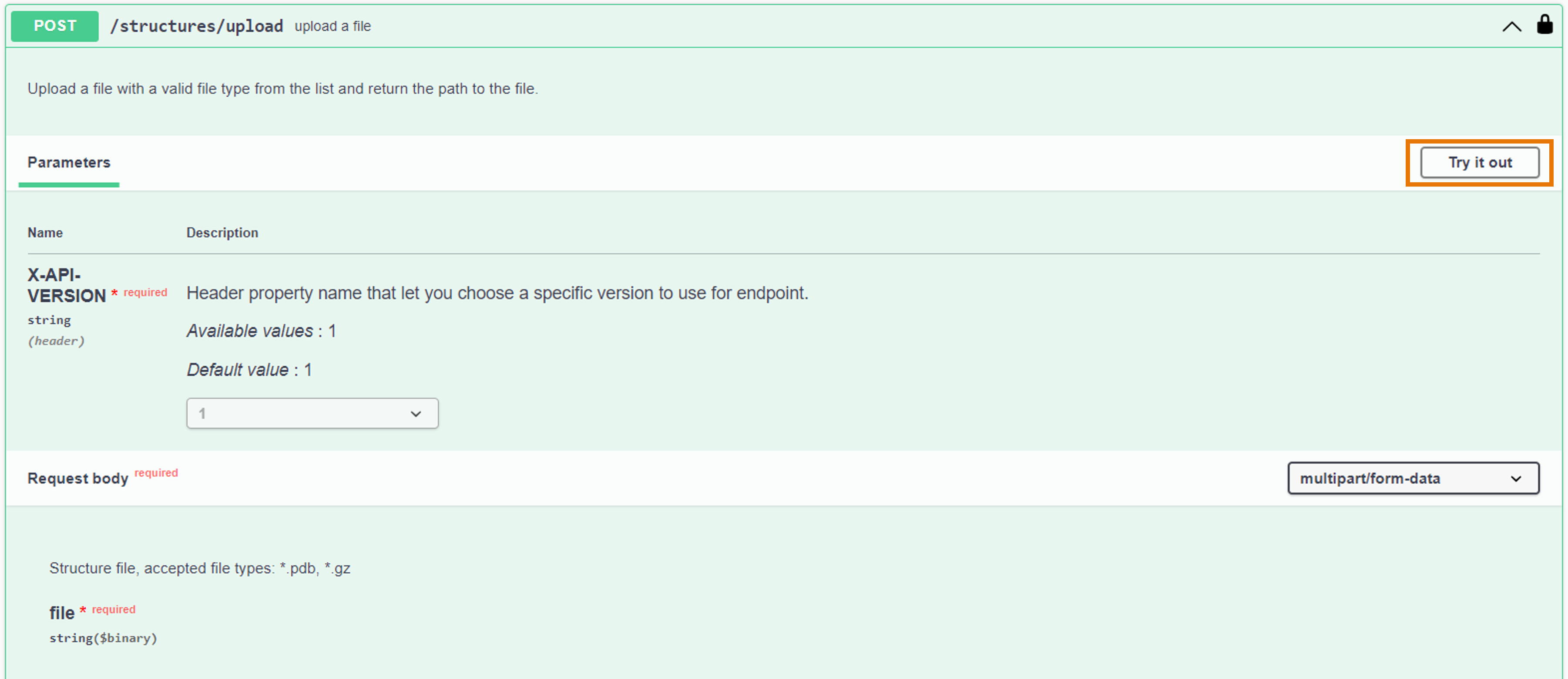
- Click on "Choose file" and add the file you want to upload
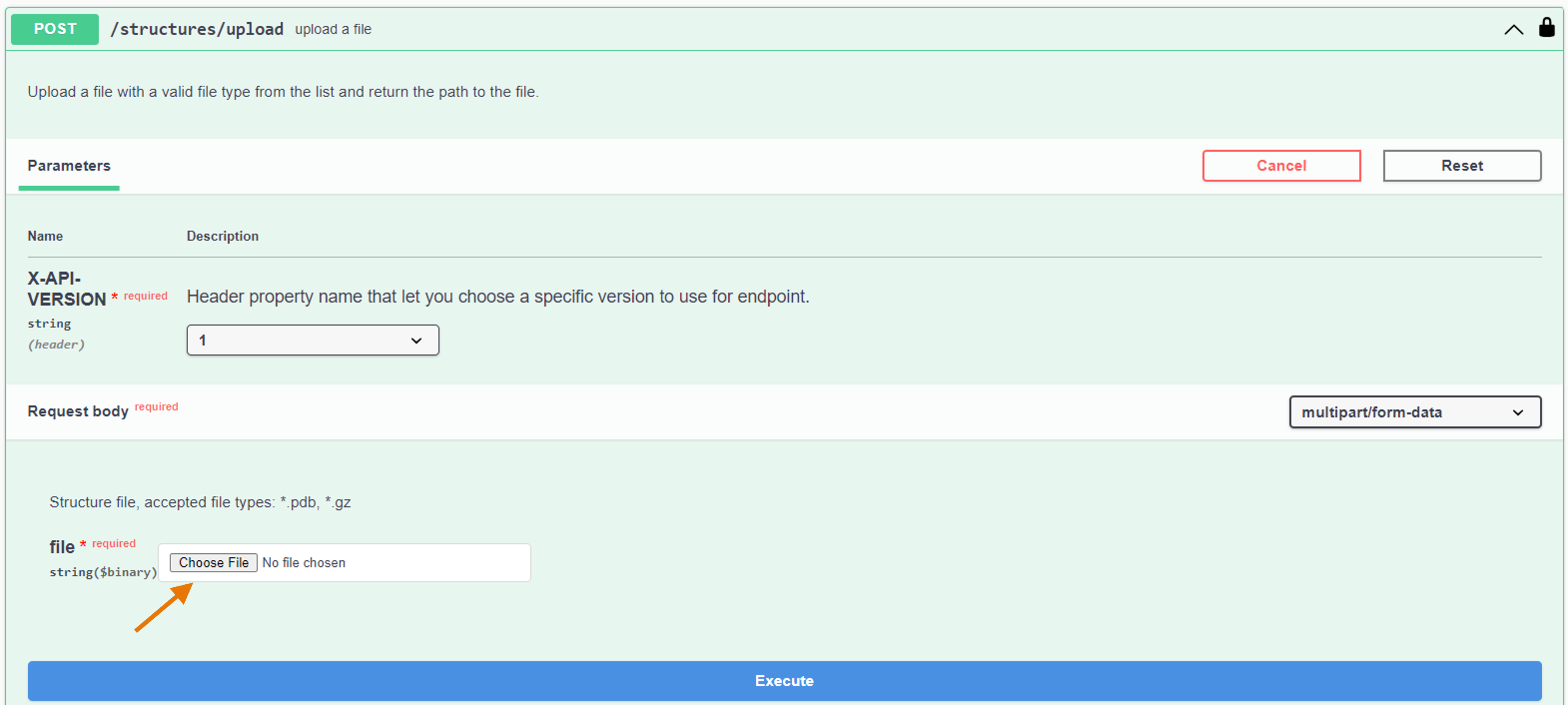
- Click on "Execute"
In the Response body, the generated server file path is reported.
¶ Using the 'curl' command
curl -X 'POST' \
'https://3decision-<customer>-api.discngine.cloud/structures/upload' \
-H 'accept: application/json' \
-H 'X-API-VERSION: 1' \
-H 'Authorization: Bearer eyJhb********XiM' \
-H 'Content-Type: multipart/form-data' \
-F 'file=@2ozo.pdb'
¶ Using Python
from pathlib import Path
import requests
# discngine_3decision_tools.py is a file that contains code to access to the API
from discngine_3decision_tools.api_utils import get_requests_session
def upload_structure(filepath: Path, session: requests.Session) -> Path:
"""
Drop a structure file on the server.
This step is required to allow further registration of a structure.
A file path on the server is returned.
Parameters
----------
filepath : Path
file path of the structure (client side)
session: requests.Session
The current session to call 3decision API.
Returns
-------
Path
file path of the structure (server side)
"""
endpoint = "https://3decision-<customer>-api.discngine.cloud/structures/upload"
# Call the upload endpoint
response = session.post(
url=endpoint,
files={"file": filepath.open("rb")},
)
if response.status_code > 399:
# Something went wrong during the endpoint call.
raise ValueError("Structure wasn't uploaded.")
# The project was successfuly added.
server_path = response.json()["path"]
return Path(server_path)
if __name__ == "__main__":
session = get_requests_session()
filepath = Path("2ozo.pdb")
server_path = upload_structure(filepath, session)
print(server_path)
¶ Response example
{
"path": "/privatedata/1/structures/upload-b6d40d5f-b758-42eb-952c-5876ccf6c233/pdb2ozo.pdb"
}
¶ 2. Submit the structure registration
The second step to register a structure is to use the POST /structure-registration endpoint.
This endpoint is asynchronous, meaning it will submit the registration as a job and return the job ID. Other endpoints can be used to check the status of the structure registration and get the job results (external code for the registered structure).
Notice that
POST /structure-registrationendpoint has a rate limit of a maximum of 1000 requests per 1000-second interval (as documented in the endpoint description in the Swagger API description).
Once the rate limit reached, calling the endpoint returns a 429 error.
¶ From the 3decision API interface
- From the activated API interface, go to the
POST /structure/registrationendpoint - Click on "Try it out" to activate the endpoint
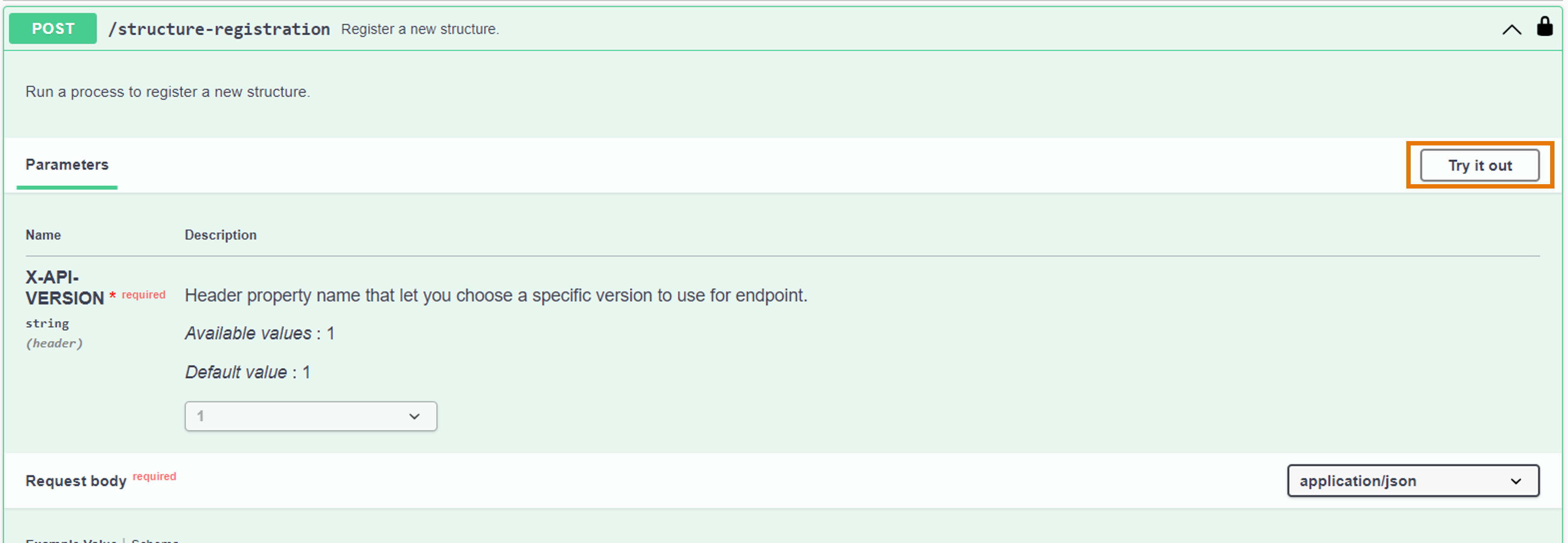
- Fill in the "Request body" with the registration payload. Here is an example you can use:
{
"experimentalMethod": "MODEL",
"commit": true,
"resolution": 0,
"filePath": "/privatedata/1/structures/upload-e5491f6a-d9f1-4fb3-a08c-0b810ec987ed/2ozo.pdb",
"title": "My last Transferase ZAP70 model",
"projectIds": [
24
],
"annotation": {
"label": "STR_564345353_model_08",
"typeLabel": "Internal ID"
},
"metadata": {
"chains": [
{
"name": "A",
"biomolCode": "ZAP70_HUMAN"
}
],
"ligands": [
{
"residueCode": "ANP",
"smiles": "c1nc(c2c(n1)n(cn2)[C@H]3[C@@H]([C@@H]([C@H](O3)CO[P@](=O)(O)O[P@@](=O)(NP(=O)(O)O)O)O)O)N",
"internalId": "ANP1"
}
]
},
"structureRelation": [
{
"externalCode": "1abcde",
"relationType": "Derived",
"hierarchyType": "Parent",
"relationDescription": "New conformation from MD"
}
]
}
In the field "filePath", use the path generated in the previous step.
- Click on
Execute
¶ Using the 'curl' command
curl -X 'POST' \
'https://3decision-<customer>-api.discngine.cloud/structure-registration' \
-H 'accept: application/json' \
-H 'X-API-VERSION: 1' \
-H 'Authorization: Bearer eyJhb********XiM' \
-H 'Content-Type: application/json' \
-d '{
"experimentalMethod": "MODEL",
"commit": true,
"resolution": 0,
"filePath": "/privatedata/1/structures/upload-e5491f6a-d9f1-4fb3-a08c-0b810ec987ed/2ozo.pdb",
"title": "My last Transferase ZAP70 model",
"projectIds": [
24
],
"annotation": {
"label": "STR_564345353_model_08",
"typeLabel": "Internal ID"
},
"metadata": {
"chains": [
{"name": "A", "biomolCode": "ZAP70_HUMAN"}
],
"ligands": [
{ "residueCode": "ANP", "smiles": "c1nc(c2c(n1)n(cn2)[C@H]3[C@@H]([C@@H]([C@H](O3)CO[P@](=O)(O)O[P@@](=O)(NP(=O)(O)O)O)O)O)N", "internalId": "ANP1"}
]
},
"structureRelation": [
{
"externalCode": "1abcde",
"relationType": "Derived",
"hierarchyType": "Parent",
"relationDescription": "New conformation from MD"
}
]
}'
¶ Using Python
import json
import requests
from discngine_3decision_tools.api_utils import get_requests_session
def submit_structure_registration_job(payload: string, session: requests.Session) -> str:
"""
Submit a structure registration Job using async structure-registration endpoint.
Parameters
----------
payload : str
all the information to register a structure
session : requests.Session
The current session to call 3decision API.
Returns
-------
str
submited registration job ID.
"""
endpoint = "https://3decision-<customer>-api.discngine.cloud/structure-registration"
response = session.post(
url=endpoint,
headers={**session.headers, **{"Content-Type": "application/json"}},
data=json.loads(payload),
)
response.raise_for_status()
# registrationId is the async job Id
return response.json()["registrationId"]
if __name__ == "__main__":
session = get_requests_session()
payload = (
'{"experimentalMethod": "MODEL","commit": true, '
'"resolution": 0, "filePath": "/privatedata/1/structures/pdb2ozo.ent.gz", '
'"title": "My last Transferase ZAP70 model", "projectIds": [24], "annotation": '
'{"label": "STR_564345353_model_08","typeLabel": "Internal ID"}, "metadata": '
'{"chains": [{"name": "A", "biomolCode": "ZAP70_HUMAN"}],"ligands": [{ '
'"residueCode": "ANP", "smiles": "c1nc(c2c(n1)n(cn2)[C@H]3[C@@H]([C@@H]'
'([C@H](O3)CO[P@](=O)(O)O[P@@](=O)(NP(=O)(O)O)O)O)O)N", "internalId": "ANP1"}]},'
'"structureRelation": [{"externalCode": "1abcde","relationType": "Derived",'
'"hierarchyType": "Parent","relationDescription": "New conformation from MD"}]}'
)
print(submit_structure_registration_job(payload, session))
¶ Response example
{
"registrationId": "01f516fb-0f2d-4a5c-a346-a00484f387bd"
}
In the response body, you are returned a registrationID.
¶ Structure Registration Status Checks
After submitting a structure registration, the job is submitted in a queue and it is processed as soon as resources are available.
When the structure registration is over, other post-registration analyses (sequence mapping, pocket detection, interaction detection, etc.) are automatically submitted.
The user can check the status of both processes - structure registration and post-registration analyses - using dedicated endpoints in the API:
- the structure registration job status with the
GET /structure-registration/job/{jobId}endpoint; - the post-registration analyses status with the
GET /structure-status/{externalCode}endpoint.
¶ 1. Check the status of the structure registration
¶ From the 3decision API interface
- From the activated API interface, go to the
GET /structure-registration/job/{jobId}endpoint - Click on "Try it out"
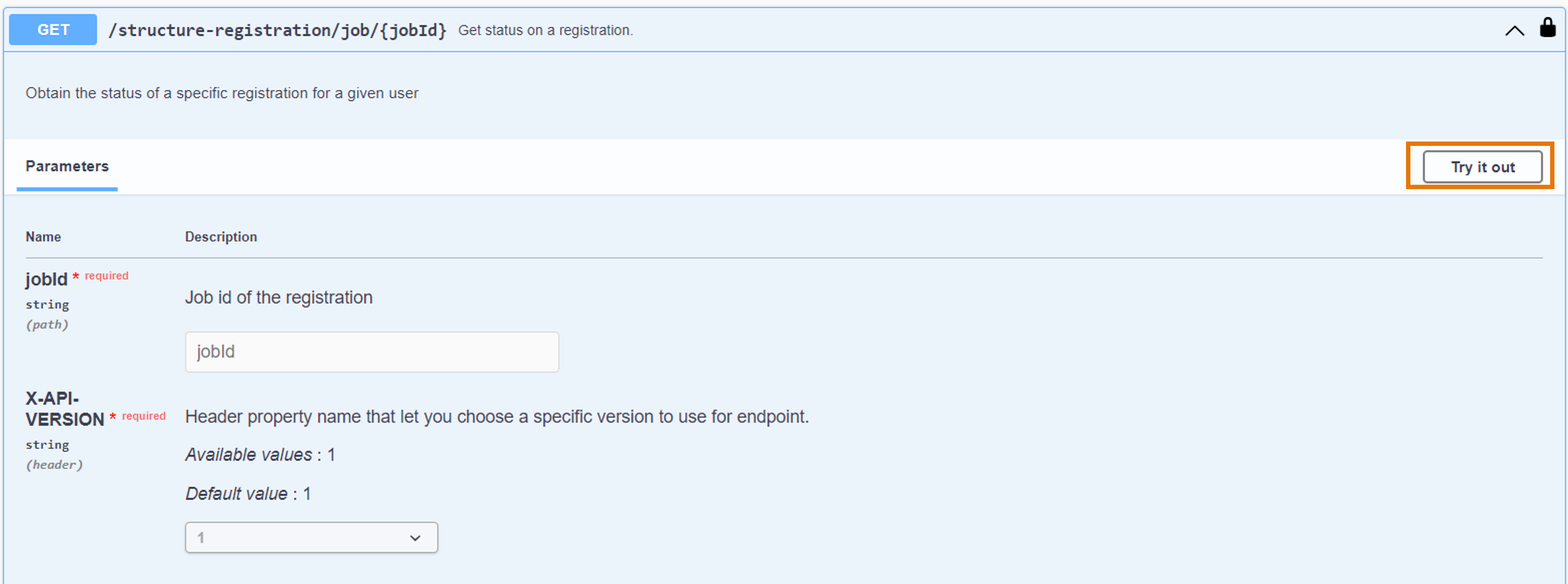
- Copy paste the job ID generated in the structure registration step
- Click on "Execute"
¶ Using the 'curl' command
curl -X 'GET' \
'https://3decision-<customer>-api.discngine.cloud/structure-registration/job/d37e520e-5cbd-4703-a03b-f411ebe79183' \
-H 'accept: application/json' \
-H 'X-API-VERSION: 1' \
-H 'Authorization: Bearer eyJhb********XiM'
¶ Using Python
from typing import Any
import requests
from discngine_3decision_tools.api_utils import get_requests_session
def get_registration_result(
registration_id: string,
session: requests.Session,
) -> dict[string, Any]:
"""
Get registration asynchronous job results.
Parameters
----------
registration_id : str
job ID of the registration process.
session : requests.Session
The current session to call 3decision API.
Returns
-------
dict[string, Any]
registered structure output.
"""
endpoint = (
"https://3decision-<customer>-api.discngine.cloud/structure-registration/job/"
+ registration_id
)
response = session.get(
url=endpoint,
headers={**session.headers, **{"Content-Type": "application/json"}},
)
response.raise_for_status()
return response.json()["content"]
if __name__ == "__main__":
session = get_requests_session()
job_id = "d37e520e-5cbd-4703-a03b-f411ebe79183"
print(get_registration_result(job_id, session))
¶ Response example
{
"status": "success",
"content": {
"externalCode": "c5twul",
"id": 784182
}
}
Once the status reaches the “success” status, the structure is available in the 3decision database. It can be accessed through both the API or the user interface using the external code
¶ 2. Check the status of the post-registration analyses
¶ From the 3decision API interface
- Go to the
GET /structure-status/{external_code}endpoint - Click on "Try it out"
- Copy paste the structure external code generated in the previous check.
- Click on "Execute"
¶ Using the 'curl' command
curl -X 'GET' \
'https://3decision-<customer>-api.discngine.cloud/structure-status/c5twul' \
-H 'accept: application/json' \
-H 'X-API-VERSION: 1' \
-H 'Authorization: Bearer eyJhb********XiM'
¶ Using Python
import json
import requests
from dng_3dec_data_import_tools.api_utils import get_requests_session
def get_structure_full_status(
external_code: string, session: requests.Session
) -> dict[string, dict[string, str] | list[dict[string, str]]]:
"""
Check all analyses and registration status.
Parameters
----------
external_code: str
the structure external code.
session : requests.Session
The current session to call 3decision API.
Returns
-------
dict[string, dict[string, str] | list[dict[string, str]]]
structure status
Raises
------
ValueError
if there is no structure status for this structure.
"""
endpoint = (
"https://3decision-<customer>-api.discngine.cloud/structure-status/"
+ external_code
)
response = session.get(url=endpoint)
json_response = json.loads(response.text)
if response.status_code > 299 or json_response.get("structureStatus") is None:
msg = "Get structure registration status failed"
raise ValueError(msg)
return json_response.get("structureStatus")
if __name__ == "__main__":
session = get_requests_session()
external_code = "c5twul"
print(get_structure_full_status(external_code, session))
¶ Response example
{
"structureStatus": {
"ligandCavityOverlapAnalysis": [
{
"stateNumber": 1,
"state": "success",
"content": [
{
"cavity_id": 33683827,
"small_mol_conf_id": 4422740
},
{
"cavity_id": 33683827,
"small_mol_conf_id": 4422739
},
{
"cavity_id": 33683853,
"small_mol_conf_id": 4422740
},
{
"cavity_id": 33683853,
"small_mol_conf_id": 4422739
}
],
"stateCount": 1,
"timeCreated": "2023-07-10T08:46:48.467+00:00",
"externalCode": "c5twul",
"domainEventType": "ligandCavityOverlapAnalysis"
}
],
"sequenceMappingAnalysis": {
"domainEventType": "sequenceMappingAnalysis",
"id": "b673ea52-7481-4d1d-bede-197e5dd34c6c",
"externalCode": "c5twul",
"userId": 3365,
"state": "success",
"content": {
"externalCode": "c5twul",
"createdBiomolStructure": 1,
"chains": [
{
"chainCode": "A",
"biomolAccession": "ZAP70_HUMAN",
"createdSeqRes": 0,
"updateStrRes": 255
},
{
"chainCode": "A",
"biomolAccession": "ZAP70_HUMAN",
"createdSeqRes": 0,
"updateStrRes": 178
},
{
"chainCode": "A",
"biomolAccession": "ZAP70_HUMAN",
"createdSeqRes": 4,
"updateStrRes": 112
}
]
},
"timeCreated": "2023-07-10T08:46:51.370+00:00"
},
"pocketDetectionAnalysis": [
{
"stateNumber": 1,
"state": "success",
"content": [
33683827,
33683828,
33683829,
33683830,
33683831,
33683832,
33683833,
33683834,
33683835,
33683836,
33683837,
33683838,
33683839,
33683840,
33683841,
33683842,
33683843,
33683844,
33683845,
33683846,
33683847,
33683848,
33683849,
33683850,
33683851,
33683852,
33683853
],
"commitSubsequentEvents": true,
"stateCount": 1,
"timeCreated": "2023-07-10T08:46:44.128+00:00",
"externalCode": "c5twul",
"domainEventType": "pocketDetectionAnalysis"
}
],
"pocketFeaturesAnalysis": [
{
"domainEventType": "pocketFeaturesAnalysis",
"id": "a33b66b2-055d-4559-8587-0e31c8170af3",
"externalCode": "c5twul",
"userId": 3365,
"stateNumber": 1,
"stateCount": 1,
"commitSubsequentEvents": false,
"state": "success",
"content": [
{
"cavity_id": 33683840,
"number_pocket_features": 5,
"number_pocket_feature_pairs": 8
},
{
"cavity_id": 33683844,
"number_pocket_features": 7,
"number_pocket_feature_pairs": 16
},
{
"cavity_id": 33683845,
"number_pocket_features": 14,
"number_pocket_feature_pairs": 33
},
{
"cavity_id": 33683846,
"number_pocket_features": 18,
"number_pocket_feature_pairs": 45
},
{
"cavity_id": 33683847,
"number_pocket_features": 26,
"number_pocket_feature_pairs": 82
},
{
"cavity_id": 33683849,
"number_pocket_features": 13,
"number_pocket_feature_pairs": 29
},
{
"cavity_id": 33683850,
"number_pocket_features": 18,
"number_pocket_feature_pairs": 60
},
{
"cavity_id": 33683851,
"number_pocket_features": 12,
"number_pocket_feature_pairs": 31
},
{
"cavity_id": 33683852,
"number_pocket_features": 8,
"number_pocket_feature_pairs": 10
},
{
"cavity_id": 33683853,
"number_pocket_features": 24,
"number_pocket_feature_pairs": 69
},
{
"cavity_id": 33683827,
"number_pocket_features": 22,
"number_pocket_feature_pairs": 63
},
{
"cavity_id": 33683828,
"number_pocket_features": 32,
"number_pocket_feature_pairs": 118
},
{
"cavity_id": 33683829,
"number_pocket_features": 15,
"number_pocket_feature_pairs": 32
},
{
"cavity_id": 33683833,
"number_pocket_features": 9,
"number_pocket_feature_pairs": 19
}
],
"timeCreated": "2023-07-10T08:46:51.646+00:00"
}
],
"interactionRegistration": [
{
"domainEventType": "interactionRegistration",
"id": "f2976cb7-855e-42ec-95a4-ac2cc0395297",
"externalCode": "c5twul",
"userId": 3365,
"stateNumber": 1,
"stateCount": 1,
"commitSubsequentEvents": false,
"state": "success",
"content": {
"interactants": 193,
"interactant_groups": 216,
"new_interactant_types": 0,
"interactant_vs_interactant_group_mappings": 284,
"interactant_group_vs_interactant_type_mappings": 631,
"new_contact_types": 0,
"new_contact_type_groups": 0,
"new_contact_type_vs_contact_type_group_mappings": 0,
"contacts": 873,
"contact_vs_contact_type_mappings": 905
},
"timeCreated": "2023-07-10T08:47:11.793+00:00"
}
],
"structureRegistration": {
"state": "success",
"content": {
"externalCode": "c5twul",
"id": 784182
},
"timeCreated": "2023-07-10T08:46:38.300+00:00",
"domainEventType": "structureRegistration",
"id": "7c269b01-47fe-488f-b242-706f6d8e55fc"
}
}
}
If all these analyses are in the "success" status, it means that the structure has been correctly registered and processed