¶ Checking PDB Structure File Compliance
To check if your PDB file is compliant with the Structure Registration process in 3decision, you can use the POST/ structure-files/validation endpoint.
This documentation outlines the potential issues in PDB files that may impact the registration of structures in 3decision, along with instructions on how to use this endpoint.
Structure files compliant with PDB standards align with 3decision Structure Registration requirements. For more information on PDB compliance, refer to the PDB documentation.
¶ PDB Structure Files compliance issues
In this section, you find a list of issues in PDB files that may affect the compliance of your PDB structure files with 3decision Structure Registration. Each issue is detailed with an indication on how to fix the problem. These issues are identified by the POST/ structure-files/validation endpoint.
The issues in the PDB Structure Files are of two types:
-
error: prevents Structure Registration. This issue makes the structure non-compliant for registration in the 3decision database. You need to fix the error to be able to register this structure.
-
warning: does not prevent Structure Registration. If you only have warnings, the structure is still considered compliant and can be registered in the 3decision database. However, there is still a hint on how to modify the file to make it also compliant with the PDB standards.
¶ Non-compliance with 3decision (validation errors)
| Name | Content | Hint |
|---|---|---|
ASCII_CHARACTERS |
PDB line contains non-ASCII characters or control ASCII characters. | Ensure that the PDB file contains only non-control ASCII characters. |
ASCII_FILE |
PDB file couldn't be read in ASCII. | Ensure that the PDB file is encoded in ASCII. |
EMPTY_ATOM_NAME |
Atom name is empty in the PDB line. | Add an atom name to this PDB line. |
EMPTY_ELEMENT_SYMBOL |
Element symbol is empty in the PDB line. | Add an element symbol to this PDB line. |
EMPTY_FILE |
PDB file is empty. | Verify if the file is not empty. |
EMPTY_LINES |
PDB file contains empty lines. | Remove empty lines from the PDB files. |
HETATM_TER_ATOM |
In the same chain, TER line can't be placed between HETATM line and ATOM line. | Remove the TER line between HETATM and ATOM lines |
INCONSISTENT_ATOM_MODELS |
PDB file contains several models which are not consistent. | Ensure all models have the same residue atoms. |
INCONSISTENT_CHAIN_MODELS |
PDB file contains several models which are not consistent. | Ensure all models have the same chains. |
NO_ATOM_NO_HETATM |
PDB file does not contain any ATOM or HETATM lines. | Ensure the PDB file contains ATOM or HETATM records. |
NO_END |
PDB file does not end with END. | Add an END record at the end of the PDB file |
RESIDUE_NUMBER_NOT_INT |
Residue number is not an integer in the PDB line. | Replace the residue number in columns 23-26 with an integer |
TER_BETWEEN_ATOMS |
A TER line can't be placed between 2 ATOM lines. | Remove the TER line between ATOM lines. |
THIRD_PARTY |
PDB file can't be parsed by required third party softwares. | This is a generic error, contact 3decision support to investigate more on the error. |
¶ Non-compliance with PDB format (validation warnings)
| Name | Content | Hint |
|---|---|---|
ATOM_NOT_AMINO_ACID |
The PDB line starts with 'ATOM' but based on its residue name, it does not correspond to an amino acid or a nucleic acid. | Substitute 'ATOM' with 'HETATM' at the beginning of the PDB line. |
ATOM_NUMBERS_NOT_SERIAL |
Atom numbers are not serial (unique and increasing). | Update atom numbers to ensure they are unique and increasing |
CHARGES |
PDB file contains charges that are not correctly formatted. | Format the charge to ensure that the symbol appears after the number. |
HETATM_AMINO_ACID |
The PDB line starts with 'HETATM' but based on its residue name, it corresponds to an amino acid or a nucleic acid. | Substitute 'HETATM' with 'ATOM' at the beginning of the PDB line. |
HOMOGLYPH |
The PDB line may contain a homoglyph (the lowercase letter 'l' is often confused with the number '1'). | Substitute letter 'l' with the upper case letter 'L' to avoid any confusion. |
LOWER_CASE_ATOMS |
PDB file contains atoms in lowercase. | Ensure the element symbol is in uppercase |
MISALIGNED_ATOM_NAME_1 |
Atom name is not right-justified starting in column 14. | The element symbol has 1 character, ensure that the atom name starts at column 14. |
MISALIGNED_ATOM_NAME_2 |
Atom name is not right-justified starting in column 13. | The element symbol has 2 characters, ensure that the atom name starts at column 13. |
NO_ATOM |
PDB contains only HETATM lines (no ATOM lines). | If the PDB file should contain a protein, check if it contains ATOM records. |
RESIDUES_OUT_OF_SEQUENCE |
Residues are out of sequence. | Ensure the residue numbers are ordered in ascending order in the same chain |
TER_AFTER_ATOM |
HETATM record found with no previous TER line. | A TER record is required and should be added if the residue is a non-polymer entity, but this warning should be ignored if the residue is a modified residue. |
TER_ATOM_NUMBERS |
TER record has no atom number. | Add an atom serial to the TER line. |
TER_BETWEEN_HETATM |
A TER line can't be placed between 2 HETATM lines. | Remove the TER line between HETATM lines. |
WRONG_ATOM_NAME |
Atom name does not start with the element symbol. | Ensure the atom name starts with the element symbol. |
¶ Note on TER lines in PDB files
In a PDB file, the presence and correct placement of TER records are crucial as they signify the end of a chain or the transition between ATOM and HETATM. Failure to include or appropriately position TER records may result in various errors or warnings within 3decision.
Here are two examples of correct TER lines placement in PDB files:
| Example 1 | Example 2 |
|---|---|
| ATOM (chain A) TER HETATM (chain A) TER ATOM (chain B) TER HETATM (chain B) END |
ATOM (chain A) TER ATOM (chain B) TER HETATM (chain A) TER HETATM (chain B) END |
¶ How to check PDB Structure File compliance
Using the 3decision API, you can check the compliance of your PDB Structure File with the 3decision Structure Registration. Also, you can get a report of the issues in the input PDB file and suggestions on how to fix them (full list in the previous section).
You first need to access and activate the API (instructions in the Access page of this documentation).
Then, a PDB Structure File can be checked in three steps:
- Upload the PDB Structure File to the 3decision server (endpoint:
POST /structures/upload) - Submit the PDB Structure File validation (endpoint:
POST /structure-files/validation) - Retrieve the compliance report on the PDB Structure File (endpoint:
GET /structure-files/validation/job/{jobId})
The instructions on how to run these enpoints are reported below. For each step, instructions from the 3decision API interface, Curl commands, and Python scripts are reported.
¶ 1. Upload the PDB Structure File
The first step to check a PDB Structure File is to upload the file to the server using the POST /structures/upload endpoint.
This endpoint will return a server file path (required for the second step).
¶ From the 3decision API interface
- From the activated API interface, go to the
POST /structures/uploadendpoint. - Click on "Try it out" to activate the endpoint.
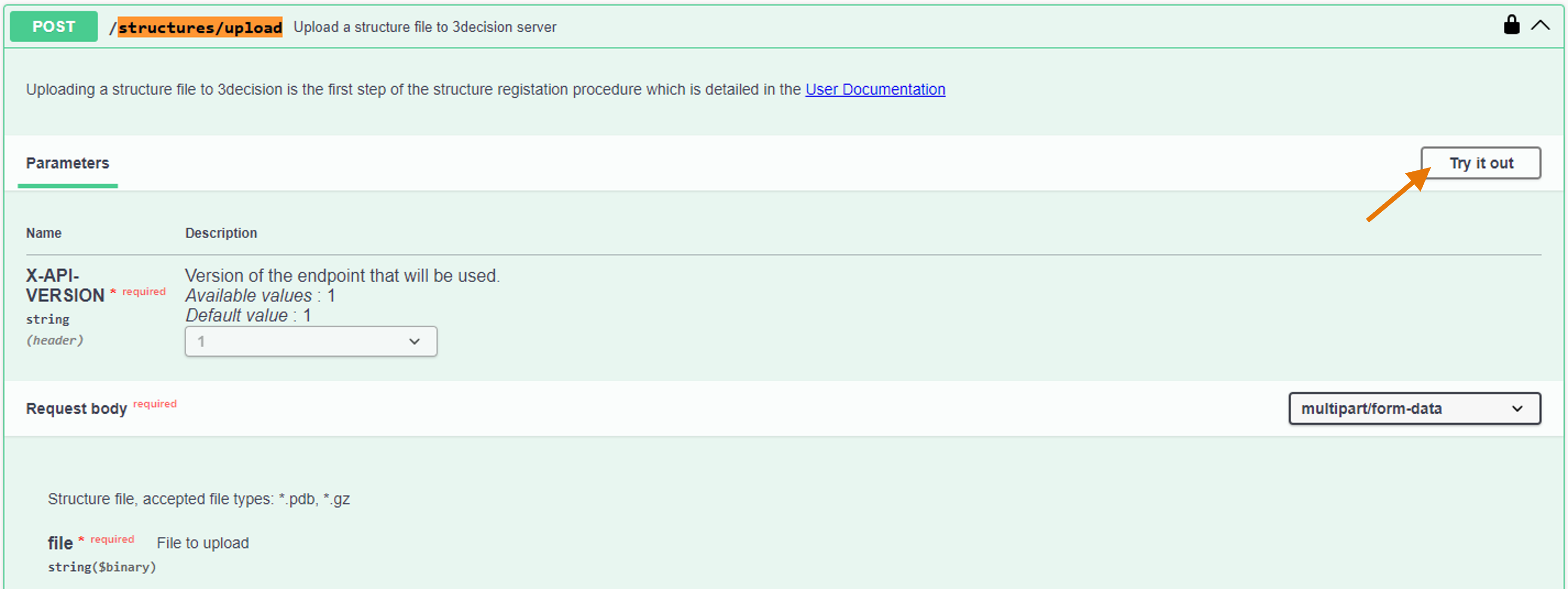
- Click on "Choose file" and add the PDB Structure File you want to check.
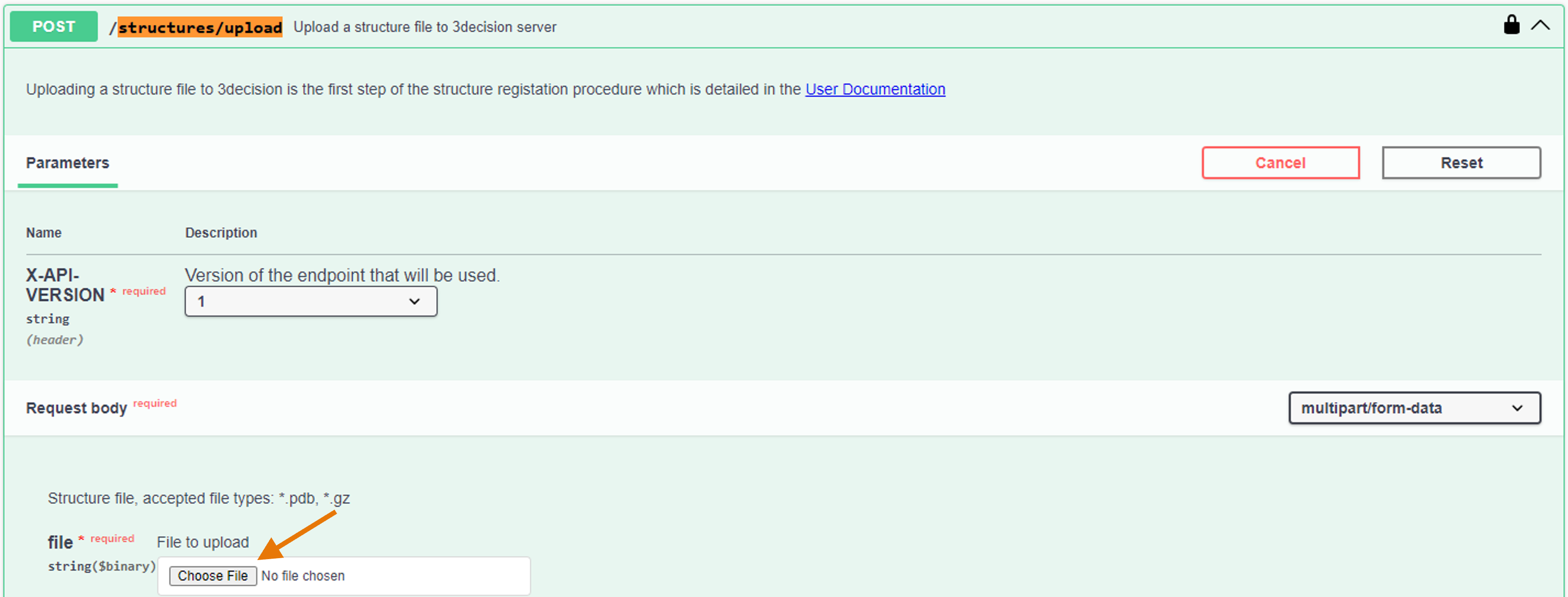
- Click on "Execute".
In the Response body, the generated server file path is reported.
¶ Using the 'curl' command
curl -X 'POST' \
'https://3decision-<customer>-api.discngine.cloud/structures/upload' \
-H 'accept: application/json' \
-H 'X-API-VERSION: 1' \
-H 'Authorization: Bearer eyJhb********XiM' \
-H 'Content-Type: multipart/form-data' \
-F 'file=@2ozo.pdb'
¶ Using Python
from pathlib import Path
import requests
# discngine_3decision_tools.py is a file that contains code to access to the API
from discngine_3decision_tools.api_utils import get_requests_session
def upload_structure(filepath: Path, session: requests.Session) -> Path:
"""
Drop a structure file on the server.
This step is required to allow further registration of a structure.
A file path on the server is returned.
Parameters
----------
filepath : Path
file path of the structure (client side)
session: requests.Session
The current session to call 3decision API.
Returns
-------
Path
file path of the structure (server side)
"""
endpoint = "https://3decision-<customer>-api.discngine.cloud/structures/upload"
# Call the upload endpoint
response = session.post(
url=endpoint,
files={"file": filepath.open("rb")},
)
if response.status_code > 399:
# Something went wrong during the endpoint call.
raise ValueError("Structure wasn't uploaded.")
# The structure was successfuly uploaded.
server_path = response.json()["path"]
return Path(server_path)
if __name__ == "__main__":
session = get_requests_session()
filepath = Path("2ozo.pdb")
server_path = upload_structure(filepath, session)
print(server_path)
¶ Response example
{
"path": "/privatedata/1/structures/upload-b6d40d5f-b758-42eb-952c-5876ccf6c233/pdb2ozo.pdb"
}
¶ 2. Submit the PDB Structure File validation
The second step to check a PDB Structure File for compliance is to use the POST /structure-file/validation endpoint.
This endpoint is asynchronous, so it will submit the validation as a job and return the job ID. This job ID is required for the third step, to retrieve the validation report.
¶ From the 3decision API interface
- From the activated API interface, go to the
POST /structure-file/validationendpoint
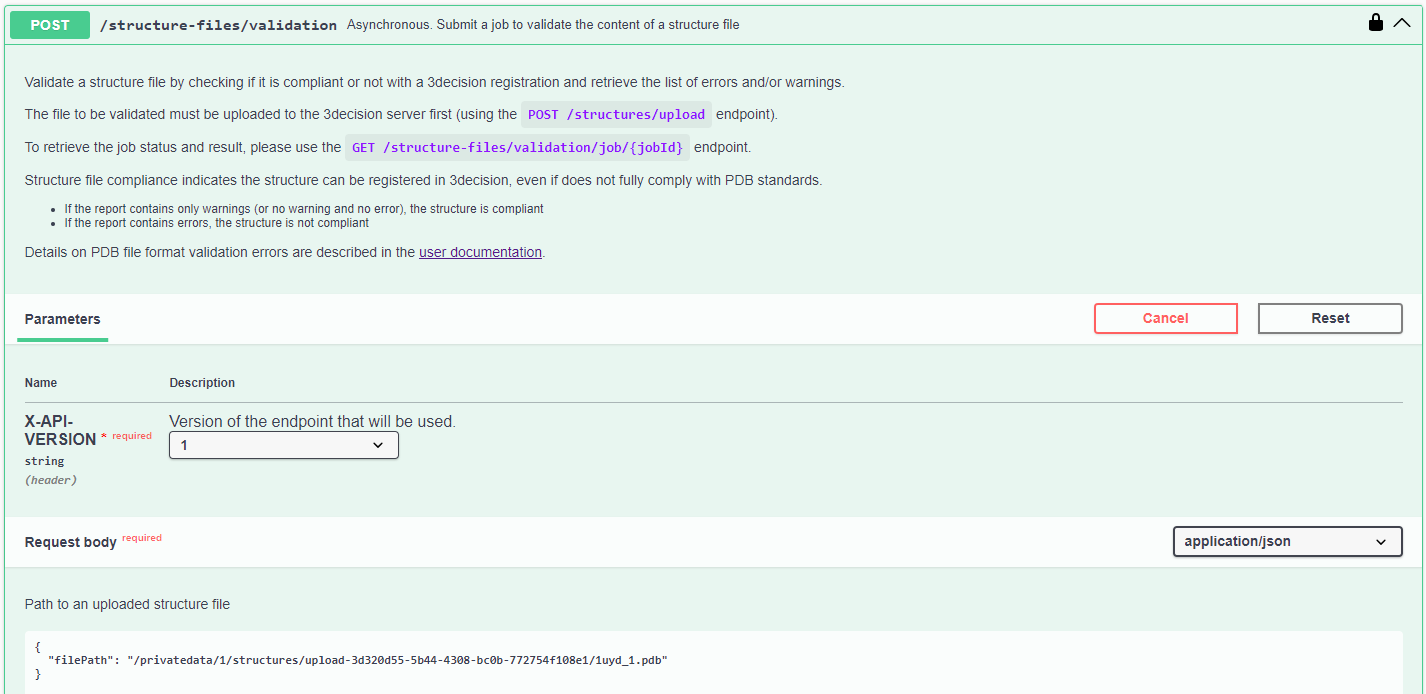
- Click on "Try it out" to activate the endpoint
- Fill in the "Request body" with the path obtained from the previous step.
- Click on
Execute
In the response body, you are returned a jobID.
¶ Using the 'curl' command
curl -X 'POST' \
'https://3decision-<customer>-api.discngine.cloud/structure-files/validation' \
-H 'accept: application/json' \
-H 'X-API-VERSION: 1' \
-H 'Authorization: Bearer eyJhb********5GA' \
-H 'Content-Type: application/json' \
-d '{
"filePath": "/privatedata/1/structures/upload-3d320d55-5b44-4308-bc0b-772754f108e1/1uyd_1.pdb"
}'
¶ Using Python
from pathlib import Path
from typing import Any
import requests
import json
# discngine_3decision_tools.py is a file that contains code to access to the API
from discngine_3decision_tools.api_utils import get_requests_session
def validate_structure_file(filepath: Path, session: requests.Session) -> str:
"""
validate a structure file
Parameters
----------
filepath : Path
Uploaded file path
session: requests.Session
The current session to call 3decision API.
Returns
-------
str
The job ID of the structure files validation job.
"""
endpoint = (
"https://3decision-<customer>-api.discngine.cloud/structure-files/validation"
)
response = session.request(
method="post",
url=endpoint,
data=str(json.dumps({"filePath": str(filepath)})),
)
if response.status_code > 399:
# Something went wrong during the endpoint call.
raise ValueError("Couldn't validate structure file.")
return response.json()["job_id"]
if __name__ == "__main__":
session = get_requests_session()
filepath = Path("2ozo.pdb")
server_path = upload_structure(filepath, session)
print(validate_structure_file(server_path, session))
¶ Response example
{
"jobId": "2e3062f8-a572-4b9a-ab2f-1cbc5a7f00ea"
}
¶ 3. Retrieve the validation report on the PDB Structure File
After submitting a PDB Structure File validation, you can check the result of the validation using the dedicated endpoint in the API: GET /structure-files/validation/job/{jobId}.
The response of this endpoint gives you the result of the PDB Structure File validation analysis. In the response content you have:
compliant: the value istruefor compliant, orfalsefor non-compliant PDB Structure Files.warnings: the value can benullis no warnings were detected, or you can have the details on the warning (see below).errors: the value can benullis no errors were detected, or you can have the details on the errors (see below).
If an error/warning is detected, the following details are provided:
name: name of the issue;content: description of the issue;hint_to_resolve: indication on how to fix the issue;line_number: the line number in the PDB file where the issue was detected;line_content: the full PDB line containing the issue;link_to_reference: a link to the PDB reference documentation.
The full list of name, content, and hint_to_resolve are provided in the first section of this page.
Examples of responses to this endpoint are reported at the end of this section.
¶ From the 3decision API interface
- From the activated API interface, go to the
GET /structure-files/validation/job/{jobId}endpoint
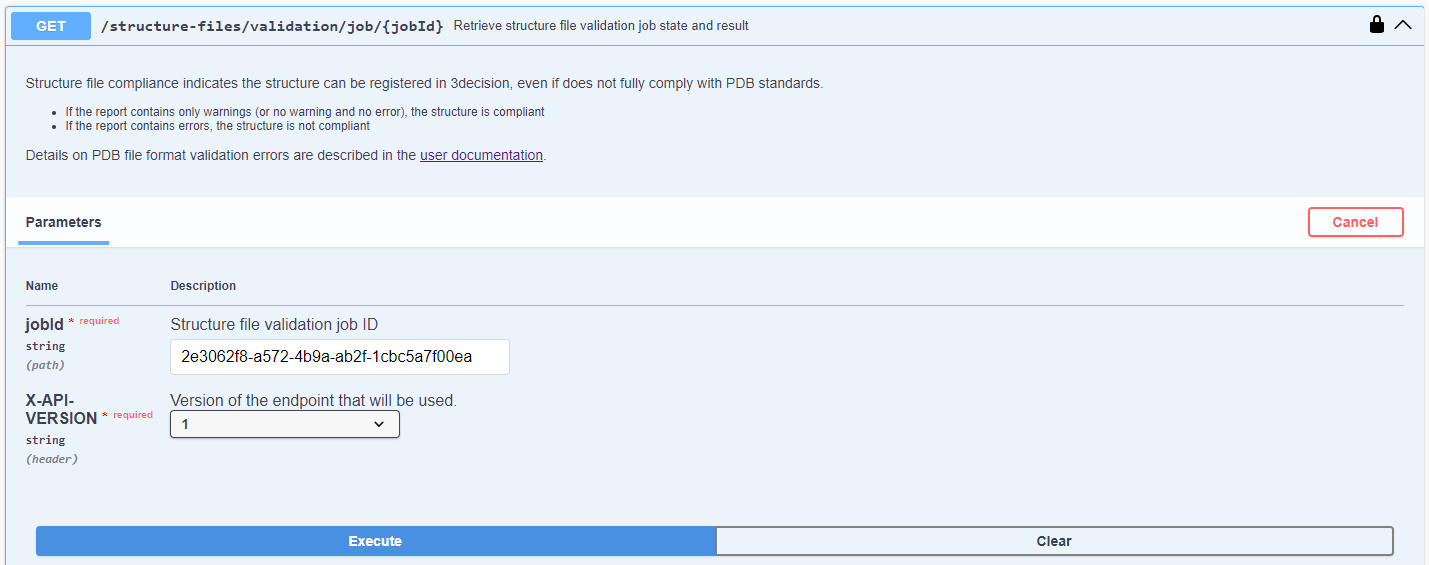
- Click on "Try it out"
- Copy paste the job ID generated in the PDB Structure File validation step
- Click on "Execute"
¶ Using the 'curl' command
curl -X 'GET' \
'https://3decision-<customer>-api.discngine.cloud/structure-files/validation/job/a50761df-9fce-4c8d-8117-819bc5151e74' \
-H 'accept: application/json' \
-H 'X-API-VERSION: 1' \
-H 'Authorization: Bearer eyJhb********XiM'
¶ Using Python
from pathlib import Path
from typing import Any
import requests
# discngine_3decision_tools.py is a file that contains code to access to the API
from discngine_3decision_tools.api_utils import get_requests_session
def get_structure_file_validation_status(job_id:str, session:requests.Session)->Any:
"""
Get the status of a structure file validation job.
Parameters
----------
job_id : str
The job ID of the structure files validation job.
session: requests.Session
The current session to call 3decision API.
Returns
-------
Any
The JSON response from the validation endpoint.
"""
endpoint = (
"https://3decision-<customer>-api.discngine.cloud/structure-files/validation/"
f"job/{job_id}"
)
return session.request(method="get", url=endpoint).json()
if __name__ == "__main__":
session = get_requests_session()
filepath = Path("2ozo.pdb")
server_path = upload_structure(filepath, session)
job_id = validate_structure_file(server_path, session)
print(get_structure_file_validation_status(job_id, session))
¶ Response examples
Fully compliant structure:
{
"content": {
"compliant": true,
"warnings": null,
"errors": null
},
"state": "success"
}
Non-complant structure:
{
"content": {
"compliant": false,
"warnings": null,
"errors": [
{
"name": "EMPTY_FILE",
"content": "PDB file is empty.",
"hint_to_resolve": "Please verify if the file is not empty.",
"line_number": null,
"line_content": null,
"link_to_reference": "https://www.wwpdb.org/documentation/file-format-content/format33/sect1.html"
}
]
},
"state": "success"
}
Once you have a compliant PDB Structure File, you can submit it to Structure Registration (read more on 3decision Structure Registration in this documentation)