With the 3decision API, you can register a custom sequence in the 3decision database. In this way, if you have proteins with non-canonical sequences, you will be able to register them. The registration of a user sequence will generate a unique Biomol_code, that can then be used in the registration of your structure, to map the novel structure on that sequence.
This section of the API documentation describes in detail how to use the appropriate endpoints to first register your user sequence, and then use it for registering a structure.
¶ Sequence Registration Input
The data required for the user sequence registration is described in the Schema of the POST /sequence-register endpoint.
The fields are detailed in the table below:
| Name ( * = required) |
Type | Example | Description |
|---|---|---|---|
proteinName* |
string |
chk1 | Name of the protein for which the new sequence will be registered. |
geneName |
string |
chek1 | Name of the gene for which the new sequence will be registered (if applicable). |
sequence* |
string |
MGNAAAAKKGSE QESVKEFLAKAK EDFLKKWESPAQ NTAHLDQFERI |
The cutomer sequence to register. Can be either a protein, peptide or DNA/RNA sequence, using a valid alphabet. |
sequenceName |
string |
My Internal Sequence Name | Name of the sequence to register. |
sequenceVersion |
number |
1 | Version of the sequence. |
sequenceComment |
string |
My sequence has been obtained using this and that | Comment describing the sequence to register. |
taxonId* |
number |
9606 | Uniprot taxon ID of the organism, which can be retrieved from UniProt. |
commit* |
boolean |
true | Commits changes to the DB. if True, the sequence is saved in database, otherwise, the registration will be a dry run: the registration will be run, but the sequence won't be persisted. |
¶ Registration of a User Sequence
To register a sequence in 3decision from the API, you first need to access and activate the API (instructions in the Access page of this documentation).
Then, the sequence can be registered from the endpoint: POST /sequence/register.
Notice that
POST /sequence/registerendpoint has a rate limit of a maximum of 50 requests per 60-second interval (as documented in the endpoint description in the Swagger API description).
Once the rate limit reached, calling the endpoint returns a 429 error.
Instructions from the 3decision API interface and Curl commands are reported in this section of the API documentation.
¶ From the 3decision API interface:
- From the activated API interface, go to the endpoint
POST /sequence/register - Click on “Try it out” to activate the endpoint
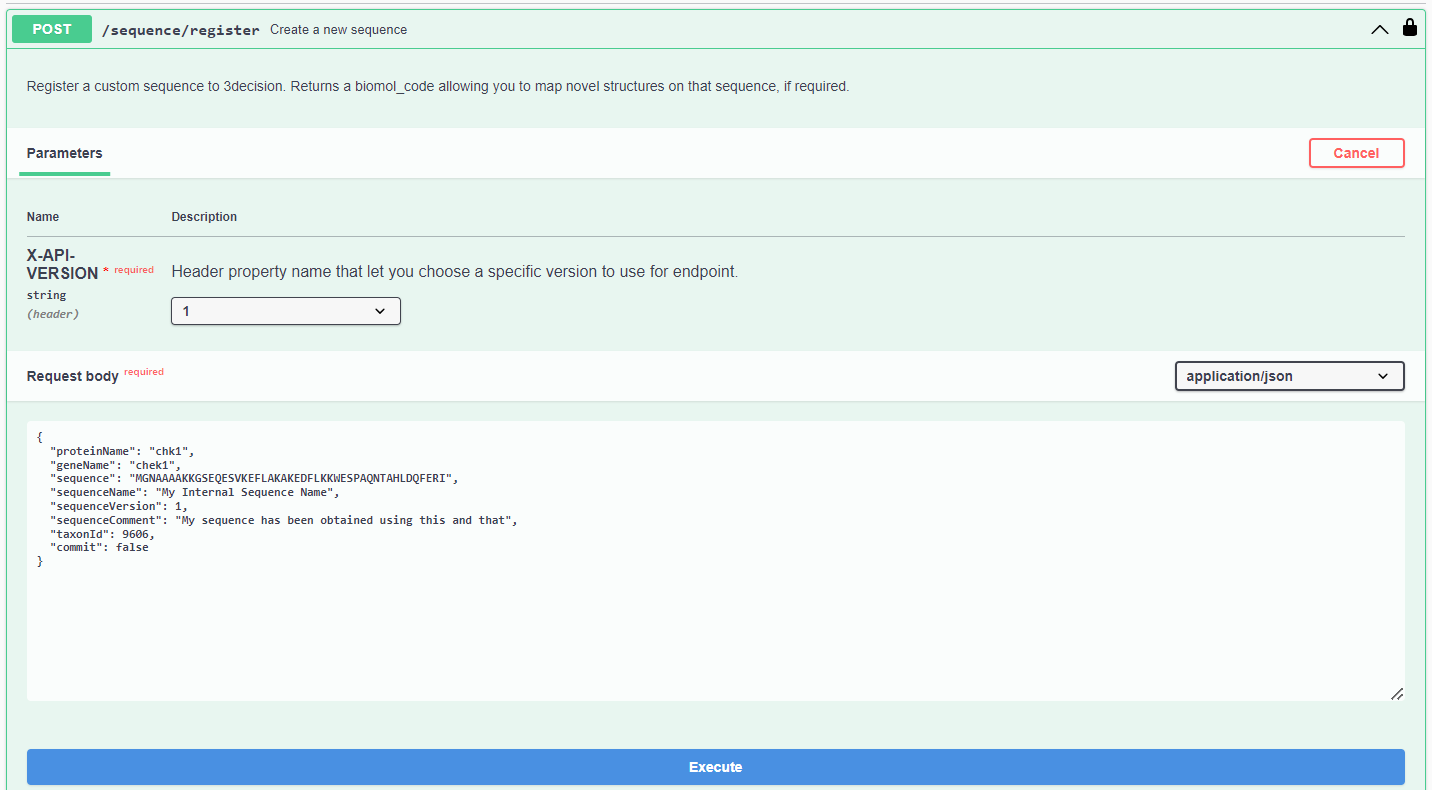
- Fill in the Request body. Here is an example that you can use:
{
"proteinName": "My Irak4",
"geneName": "IRAK4",
"sequence": "MNKPITPSTYVRCLNVGLIRKLSDFIDPQEGWKKLAVAIKKPSGDDRYNQFHIRRFEALLQTGKSPTSELLFDWGTTNCTVGDLVDLLIQNEFFAPASLLLPDAVPKTANTLPSKEAITVQQKQMPFCDKDRTLMTPVQNLEQSYMPPDSSSPENKSLEVSDTRFHSFSFYELKNVTNNFDERPISVGGNKMGEGGFGVVYKGYVNNTTVAVKKLAAMVDITTEELKQQFDQEIKVMAKCQHENLVELLGFSSDGDDLCLVYVYMPNGSLLDRLSCLDGTPPLSWHMRCKIAQGAANGINFLHENHHIHRDIKSANILLDEAFTAKISDFGLARASEKFAQTVMTSRIVGTTAYMAPEALRGEITPKSDIYSFGVVLLEIITGLPAVDEHREPQLLLDIKEEIEDEEKTIEDYIDKKMNDADSTSVEAMYSVASQCLHEKKNKRPDIKKVQQLLQEMTAS",
"sequenceName": "My Irak4 Sequence Name",
"sequenceVersion": 1,
"sequenceComment": "My custom Irak4 sequence",
"taxonId": 37438,
"commit": true
}
- Click on “Execute”
¶ Using the 'curl' command:
curl -X 'POST' \
'https://3decision-<customer>-api.discngine.cloud/sequence/register' \
-H 'accept: application/json' \
-H 'X-API-VERSION: 1' \
-H 'Authorization: Bearer eyJhb********htE' \
-H 'Content-Type: application/json' \
-d '{
"proteinName": "My Irak4",
"geneName": "IRAK4",
"sequence": "MNKPITPSTYVRCLNVGLIRKLSDFIDPQEGWKKLAVAIKKPSGDDRYNQFHIRRFEALLQTGKSPTSELLFDWGTTNCTVGDLVDLLIQNEFFAPASLLLPDAVPKTANTLPSKEAITVQQKQMPFCDKDRTLMTPVQNLEQSYMPPDSSSPENKSLEVSDTRFHSFSFYELKNVTNNFDERPISVGGNKMGEGGFGVVYKGYVNNTTVAVKKLAAMVDITTEELKQQFDQEIKVMAKCQHENLVELLGFSSDGDDLCLVYVYMPNGSLLDRLSCLDGTPPLSWHMRCKIAQGAANGINFLHENHHIHRDIKSANILLDEAFTAKISDFGLARASEKFAQTVMTSRIVGTTAYMAPEALRGEITPKSDIYSFGVVLLEIITGLPAVDEHREPQLLLDIKEEIEDEEKTIEDYIDKKMNDADSTSVEAMYSVASQCLHEKKNKRPDIKKVQQLLQEMTAS",
"sequenceName": "My Irak4 Sequence Name",
"sequenceVersion": 1,
"sequenceComment": "My custom Irak4 sequence",
"taxonId": 37438,
"commit": true
}'
¶ Response example
{
"biomolecule": {
"proteinPrimaryName": "My Irak4",
"genePrimaryName": "IRAK4",
"organism": {
"name": "Malacothrix typica",
"id": 276647,
"taxonId": 37438,
"organismId": 276647,
"organismLabel": "Malacothrix typica",
"organismCode": 37438
},
"entryVersion": 1,
"biomoleculeId": 11161653,
"sourceId": 9003,
"biomoleculeCode": "PRO_ZXHEAZ",
"type": "Protein"
},
"biomoleculeSequenceId": 11238612,
"isCanonical": "1",
"sequenceVersion": 1,
"sequence": "MNKPITPSTYVRCLNVGLIRKLSDFIDPQEGWKKLAVAIKKPSGDDRYNQFHIRRFEALLQTGKSPTSELLFDWGTTNCTVGDLVDLLIQNEFFAPASLLLPDAVPKTANTLPSKEAITVQQKQMPFCDKDRTLMTPVQNLEQSYMPPDSSSPENKSLEVSDTRFHSFSFYELKNVTNNFDERPISVGGNKMGEGGFGVVYKGYVNNTTVAVKKLAAMVDITTEELKQQFDQEIKVMAKCQHENLVELLGFSSDGDDLCLVYVYMPNGSLLDRLSCLDGTPPLSWHMRCKIAQGAANGINFLHENHHIHRDIKSANILLDEAFTAKISDFGLARASEKFAQTVMTSRIVGTTAYMAPEALRGEITPKSDIYSFGVVLLEIITGLPAVDEHREPQLLLDIKEEIEDEEKTIEDYIDKKMNDADSTSVEAMYSVASQCLHEKKNKRPDIKKVQQLLQEMTAS",
"sequenceLength": 460,
"biomoleculeSequenceName": "My Irak4 Sequence Name",
"biomolSequenceComment": "My custom Irak4 sequence"
}
Make sure that the
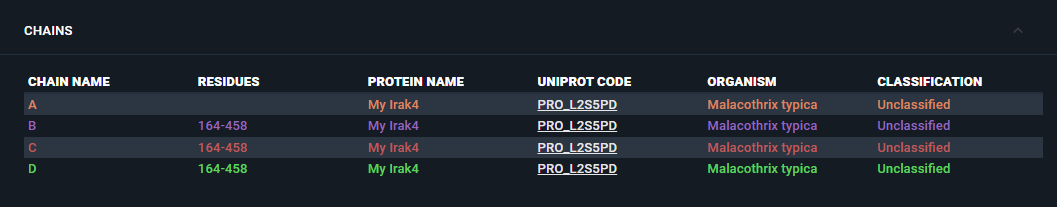
biomoleculeCodeis there: you will need it for the registration of your structure with the customed sequence. In this example, for instance, the Biomolecule Code to use for the structure registration isPRO_ZXHEAZ.
¶ Register a structure with customized User Sequence
To register a structure with the newly registered User Sequence, you can proceed as any other Structure registration (for details and step-by-step guide, refer to the Structure Registration section of this API documentation).
The only difference is that in the payload you will provide in the POST /structure-registration endpoint, you should be careful to insert the biomoleculeCode generated upon sequence registration, in the chains identifiers.
For the sequence registered in this example, your payload should look like this:
{
"experimentalMethod": "X-RAY DIFFRACTION",
"commit": true,
"resolution": 2.00,
"filePath": "/privatedata/1/structures/upload-4b2c35a4-0a32-4261-8783-804e707699d0/2nru.pdb",
"title": "API documentation example",
"projectIds": [],
"metadata": {
"chains": [
{
"name": "A",
"biomolCode": "PRO_ZXHEAZ"
},
{
"name": "B",
"biomolCode": "PRO_ZXHEAZ"
},
{
"name": "C",
"biomolCode": "PRO_ZXHEAZ"
},
{
"name": "D",
"biomolCode": "PRO_ZXHEAZ"
}
],
"ligands": []
},
"structureRelation": []
}
Your structure with the newly registered User Sequence should be available in the 3decision UI.
If you open the "Information Browser" for the protein, under the "Chains" section, you will see the full list of registered chains, with the Biomolecule Code, Protein Name and Organism, provided upon registration of the Custom Sequence.