¶ Introduction
In 3decision, you have various ways to search for structures and their associated ligands. Search results can be examined using different views. You can refine your initial search results through the use of filters or subqueries.
At the center of the 3decision Homepage, you can quickly access all the search options:
- Basic Search
- Advanced Search
- Chemistry Search

In this section, we will go through all the 3decision search options, and the results navigation page.
¶ Basic Search
You can quickly start a search by typing text in the Basic Search bar on the Homepage.
3decision searches across multiple fields associated with a structure, including compound IDs, protein or gene names, PDB codes, and more.
You can also paste a SMILES string to find a molecule or similar molecules. The similarity search uses Tanimoto similarity with a 0.7 cutoff, ensuring results are structurally close to your input.
The basic search algorithm was redesigned in 3decision 2.2.0 to deliver faster performance and more relevant results. Below are the key improvements.
¶ In versions prior to 2.2
The basic text search is rather permissive and allows to some extent misspelled names.
The following fields are covered by the basic search:
- Structure code (i.e. PDB code or '6-character 3decision code')
- Gene code
- Gene name
- UniProt accession code (e.g. P00519)
- UniProt entry name (e.g. ABL1_HUMAN)
- 3decision biomolecule codes (e.g. PRO_Q0HSLB)
- Structure title
- Protein name
- Structure annotations
- Small molecule annotations (e.g. internal compound ID)
¶ What’s New in 2.2
The Basic Search feature has been significantly enhanced in this release, improving both relevance and performance:
¶ Search Logic & Matching
- Precise Matching: Searches now rely on exact matches within the list of searchable fields, which improves speed and performance. Note that this change reduces tolerance for misspellings.
- Improved Compound Number Recognition: Searches for compound numbers in titles are now more accurate. For example, a query for
2453436will correctly match titles containingD2453436, which was previously unsupported.
¶ Expanded Searchable Fields
- Project Names Added: Basic search now includes project names as part of the searchable fields.
¶ Relevance Scoring
- Smarter Ranking: Results are now ranked by relevance, ensuring the most pertinent entries appear first (see diagram below).
¶ Basic Search Diagram
¶ Search results
When you do a search, this is how 3decision shows results.
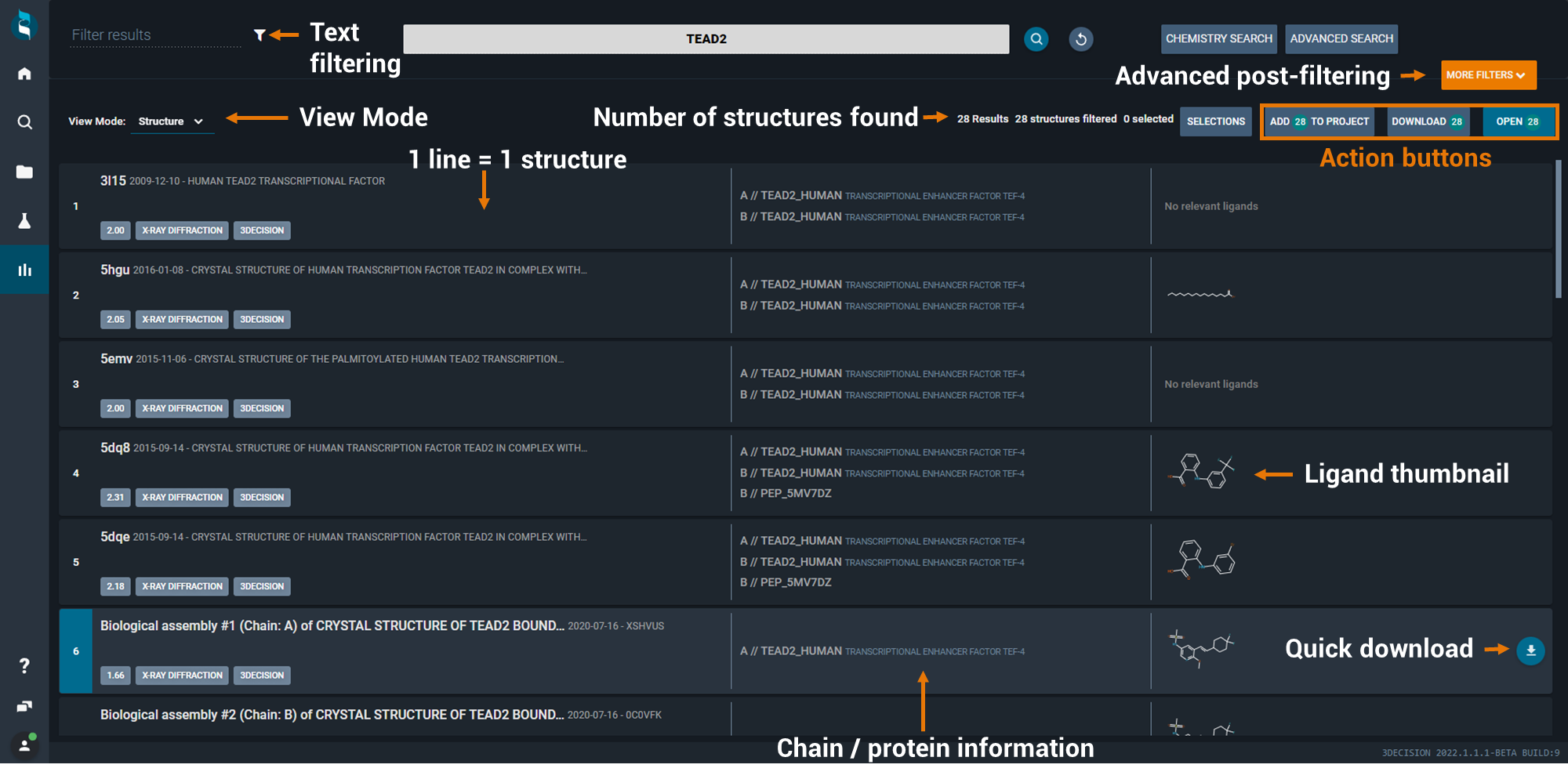
From a search results page you can basically undertake 5 main actions:
- Post-filter the search results
- Open structures within the 3decision Workspace
- Download structures or information associated to the structures
- Add structures to a project
- Start a new search
In order to open, add or download (all or only selected) structures, 3 buttons are available on the top right of the screen. If you only want to act on a subset of hit structures, you can select them by clicking (single left mouse click) on individual result lines.
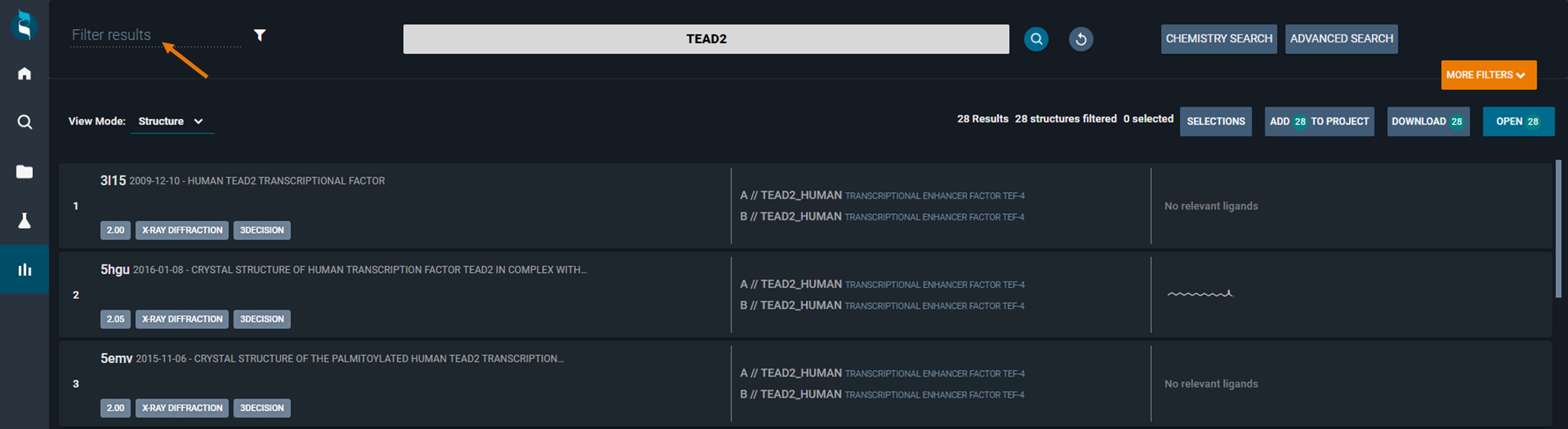
¶ View Modes
3decision allows you to visualize the result list with different view modes based on the information is more relevant to you. You can switch the view mode by clicking on the combo-box in the top-left corner of the screen.
You have the following options to choose from:
- Structure
- Ligand
- Target
- Table
¶ Structure View Mode
Each structure identified during the search is shown on a single line highlighting the structure identifier, deposition date of the structure, title, the protein(s) (Uniprot code(s)) and if there's a ligand, small molecule depiction. If your user profile is either "Crystallographer" or "Molecular Modeller", then your search results will be by default shown with this viewing mode.
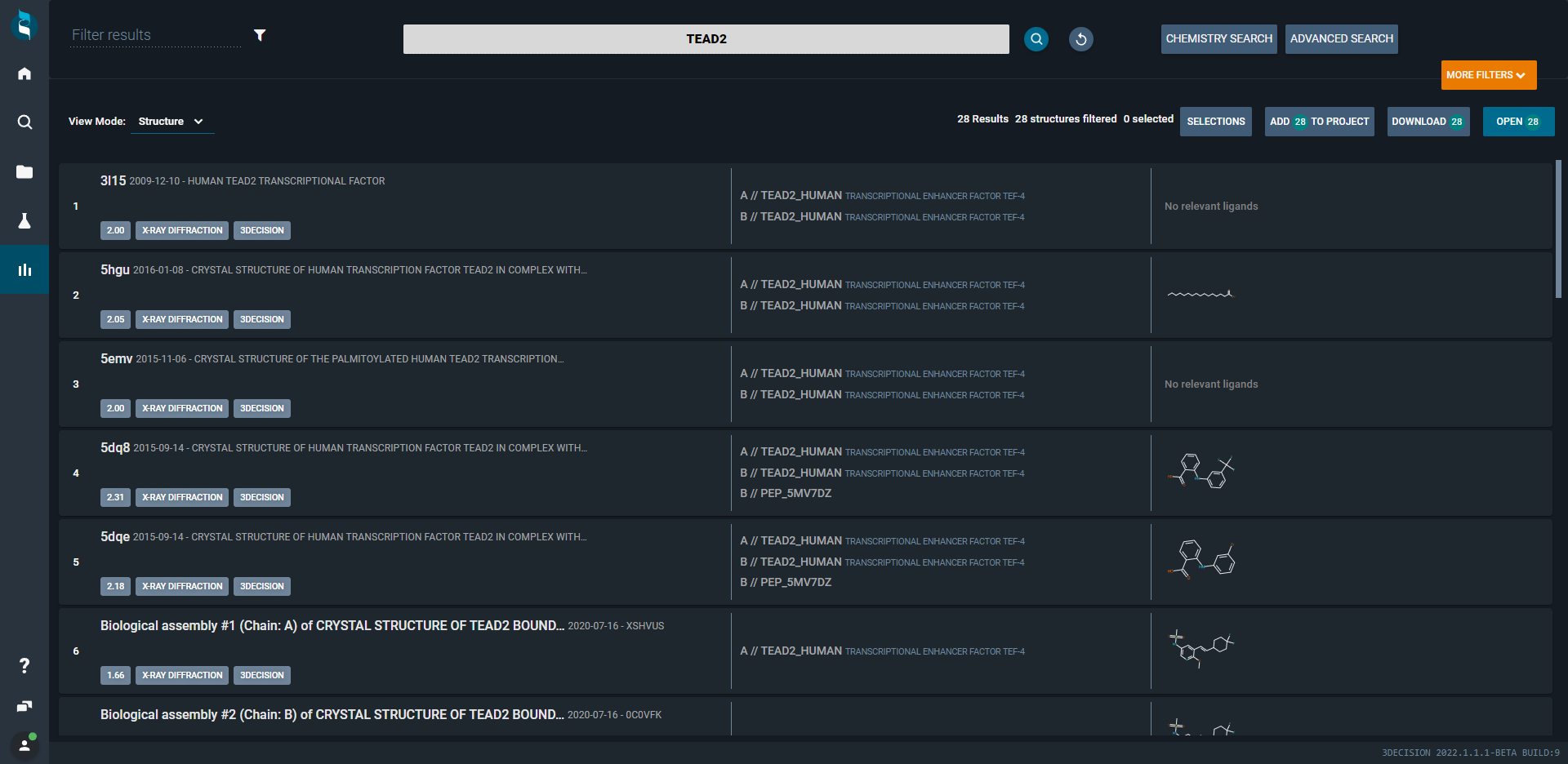
¶ Ligand View Mode
Each structure is shown as a tile in a gallery view mode focusing on what is considered to be the main ligand contained in a structure.
If your user profile is "Medicinal Chemist' then your search results will be by default shown with this viewing mode.
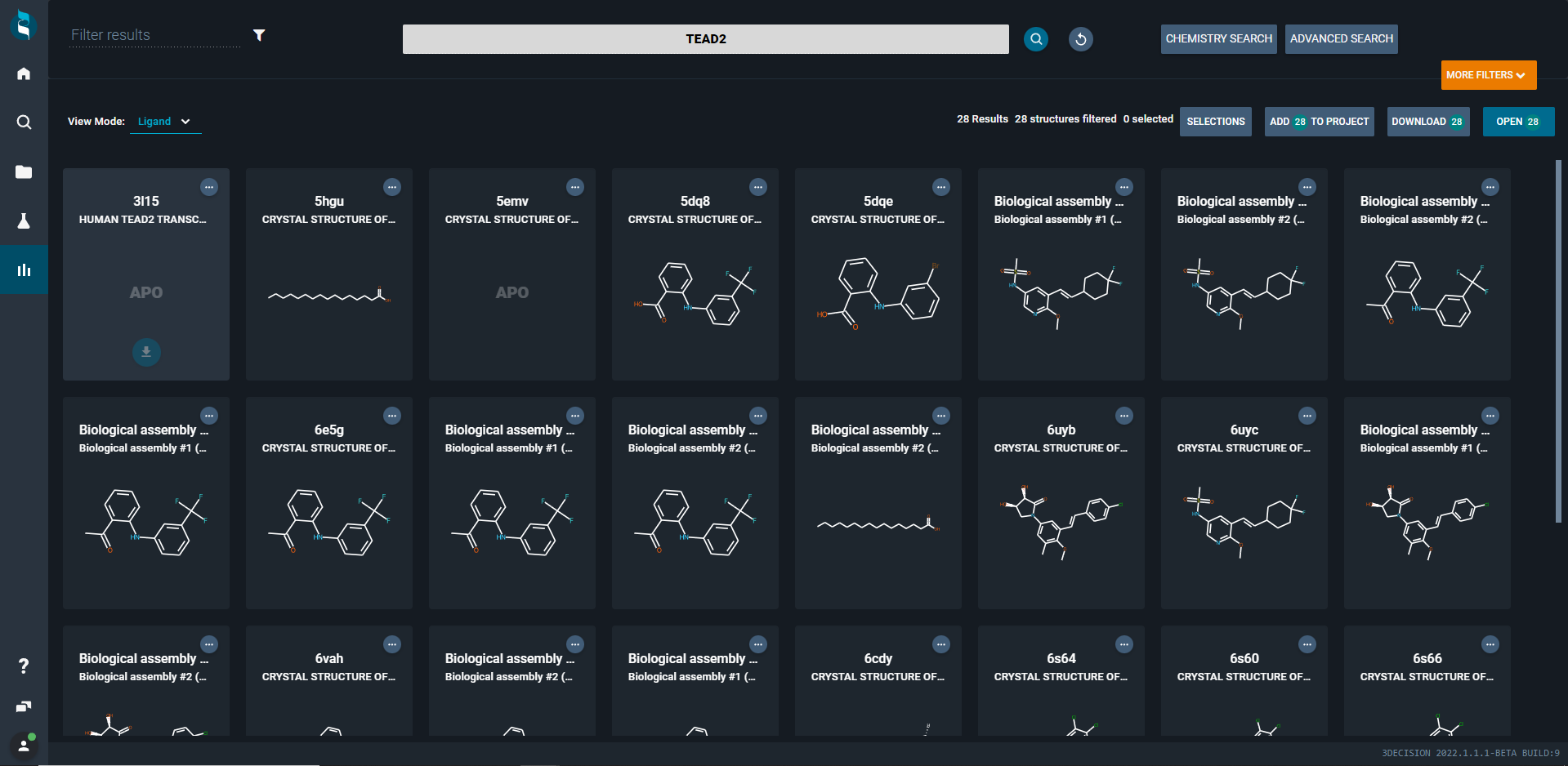
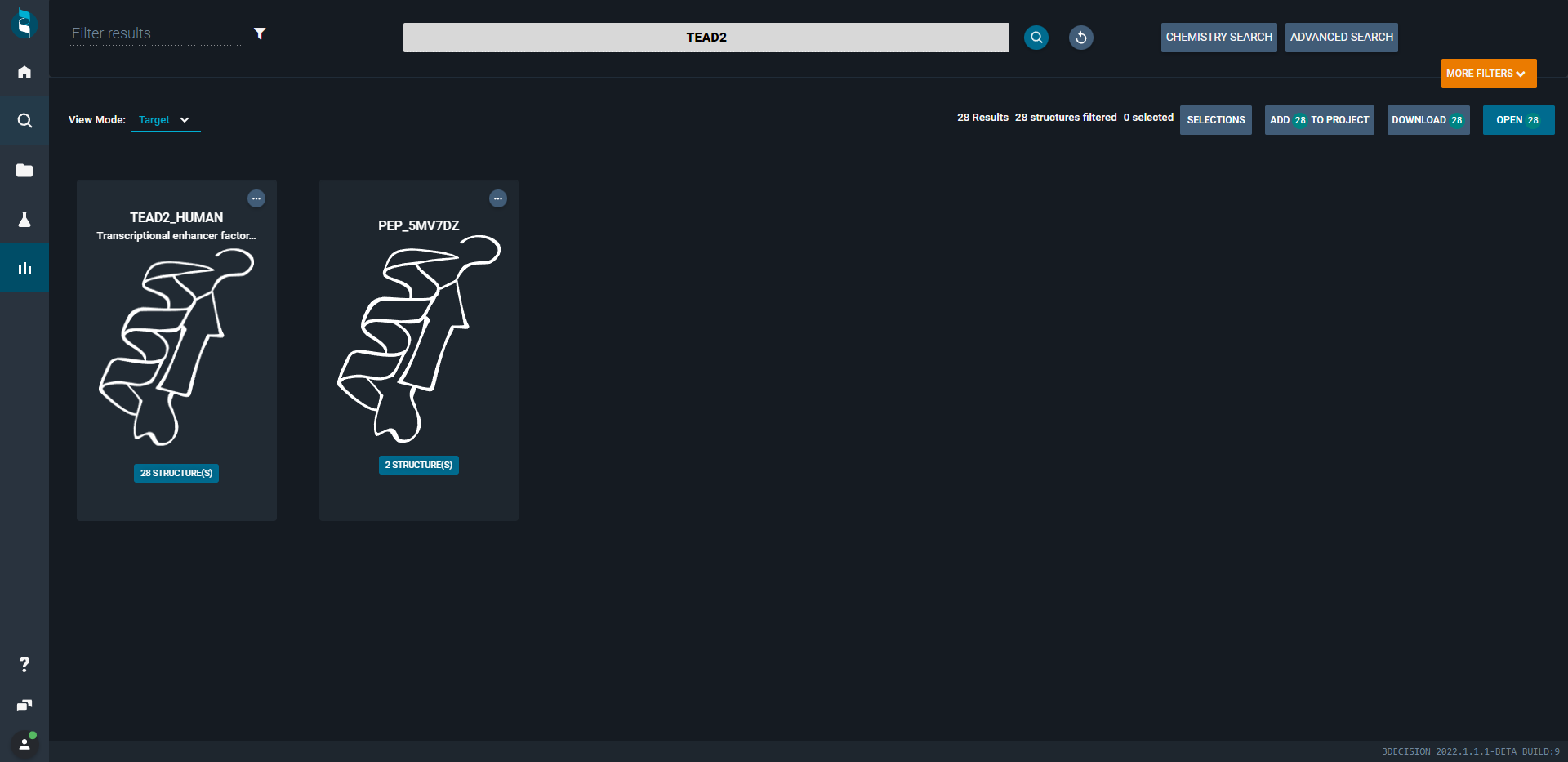
¶ Target View Mode
If you want to explore what targets (biomolecules) a search hits rather than showing individual structures, you can open the target view mode. This will collapse all structures underneath target tiles. It gives you a quick overview how many and which targets you retrieved with your search.
¶ Table View Mode
This view mode allows you to show, sort, and select structures (one line for each structure) based on several descriptors and metadata of the structures. The table collects detailed information on the listed structures, but it can be customazed to view only the columns you are interested in, using the Headers drop-down menu on the right above the table. From this menu, you can toggle on and off all columns at the same time using Toggle all. Below, some of the descriptors contained in the table are better described:
- External Code: unique identifier for each structure registered in the database. For public structure, it is the 4-letter PDB code; for internal structures, it is a 6 letter code.
- IMPORTED_BY: who imported the structure. For public structures, 'automatic_import' or '3decision' will be indicated; for proprietary structures,the user who imported the structure will be indicated.
- LIGANDS-MOLECULE_INCHI_AUX_INFO: is an InChI representation of the molecule enhanced with 3D coordinates.
- LIGANDS - STR_RESIDUE_NUMBER: is the residue number of the ligand in the structure.
- LIGANDS - ISDRUGLIKE: ligand is druglike if it has Molecular Weight between 150 and 2000, has at least one ring, and has a single bond fraction < 0.95 (single bond fraction = # single bonds / total # of bonds in the molecule).
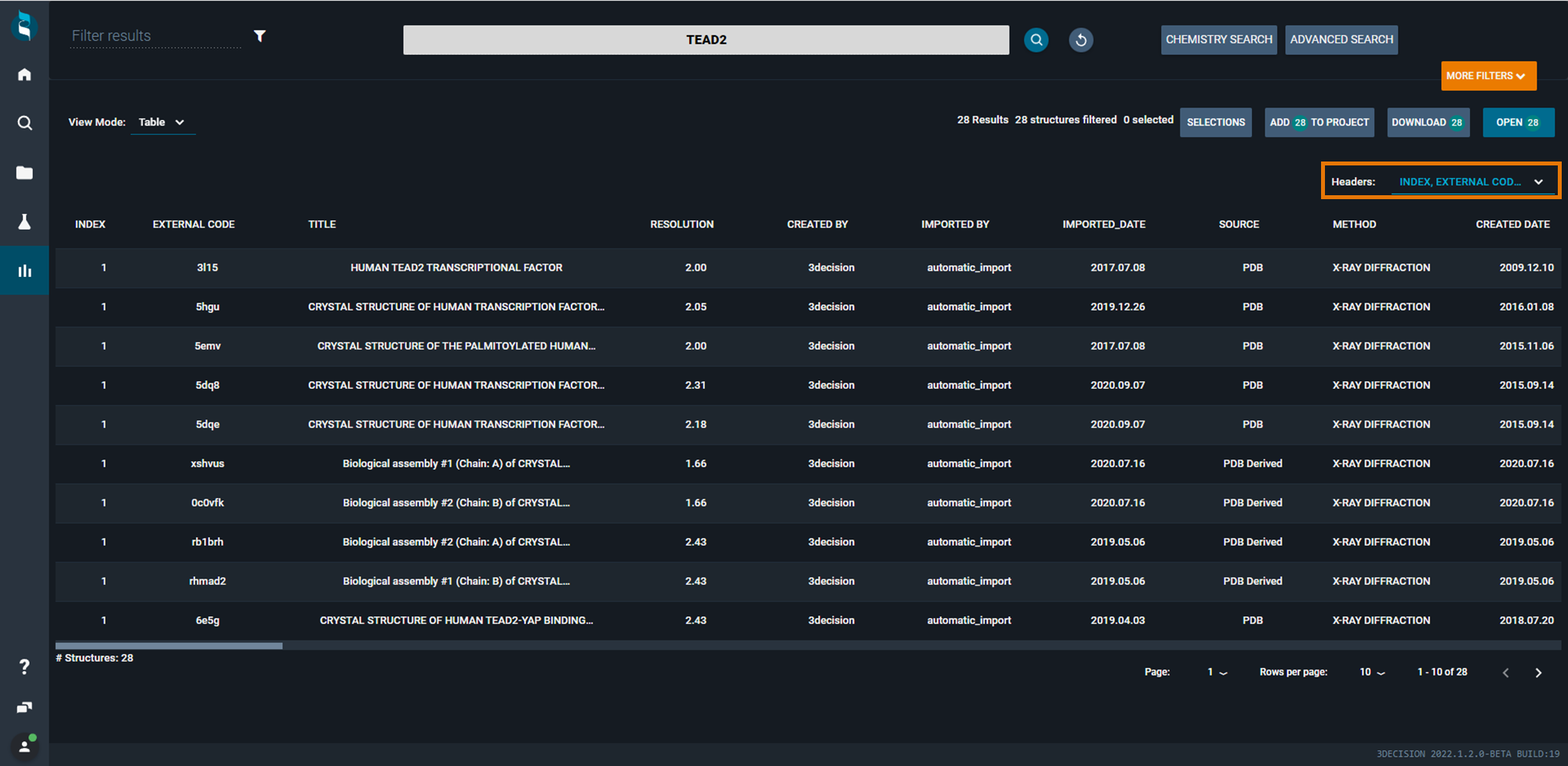
You can extract detailed information on the structures retrieved in the search, by downloading the table as it is (using the Download button at the top right of the window).
As in other view modes, you can select individual rows to either open them, add structures to a project or download data from these selected structures.
¶ Post Filtering
If you want to further refine your list of results, you can post-filter your search results within the search results page.
On the top-left corner, you have a text field that allows you to run basic text matching within all your search results. Type "ENTER" to reduce the list of shown structures to only structures containing the keyword you wrote.
Instead of running pure text searches, you can also adjust search results using more advanced post-filtering rules. In order to access these, click on the orange button MORE FILTERS
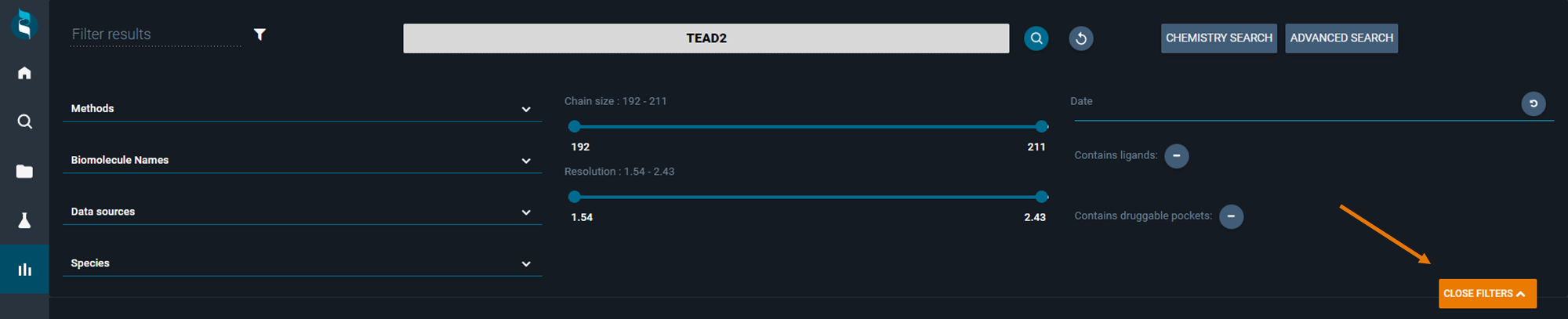
This opens up a small menu in the top of the search results page allowing you to post-filter your structures by:
- Methods: the experimental method used to solve the structure (multiple checkboxes)
- Biomolecule Names: the name of the protein (multiple checkboxes)
- Data Sources: the source where the structure is coming from (in-house, public domain, etc)
- Species: organism from which the protein belongs
- Chain Size: the number of amino acids in a structure (chain). Helpful when you look for peptide structures for instance
- Resolution: experimental resolution for structures
- Date: deposition dates
- Contains ligands: filter either structure containing or not small molecules
- Contains druggable pockets: filter structures where at least one druggable pocket has been identified (fpocket)
¶ Actions on search results
¶ Open in 3decision
Open all structures or only the selected ones (if you selected some specific structures) in the 3decision Workspace.
At any time, when within the workspace you can go back to your search results using the toolbar icon on the left:
¶ Download
Instead of opening structures you searched for, you can also download these structures or associated metadata directly from the search results page.
When clicking this button you'll be able to download the following formats/data:
- All information about structures in csv or xlsx file
- A zip file containing all structures in PDB format
- All information about ligands in xlsx or csv file
- All ligands as sdf format
¶ Add to project
In case you already set up a project before, you can add either all or a selection of your structures from your search results directly to your project(s) of interest. If you have not created a project yet, you can do that directly within the search results page. Simply type the project name you'd like to create and 3decision will suggest you to create a new project.
For information on how to create projects see Projects.
¶ Advanced Search
In the 3decision search bar, you can search from a lot of different information, but it's limited to a single string or number. Sometimes you might want to find structures according to a more specific search pattern.
For example, you want to identify all structures of a particular protein family containing a particular substructure in ligands that are cocrystallized and have a good resolution. In order to undertake these types of searches in 3decision, you should click on the magnifying glass in the main menu to open up the Advanced Search page.
The advanced search page will look like this:
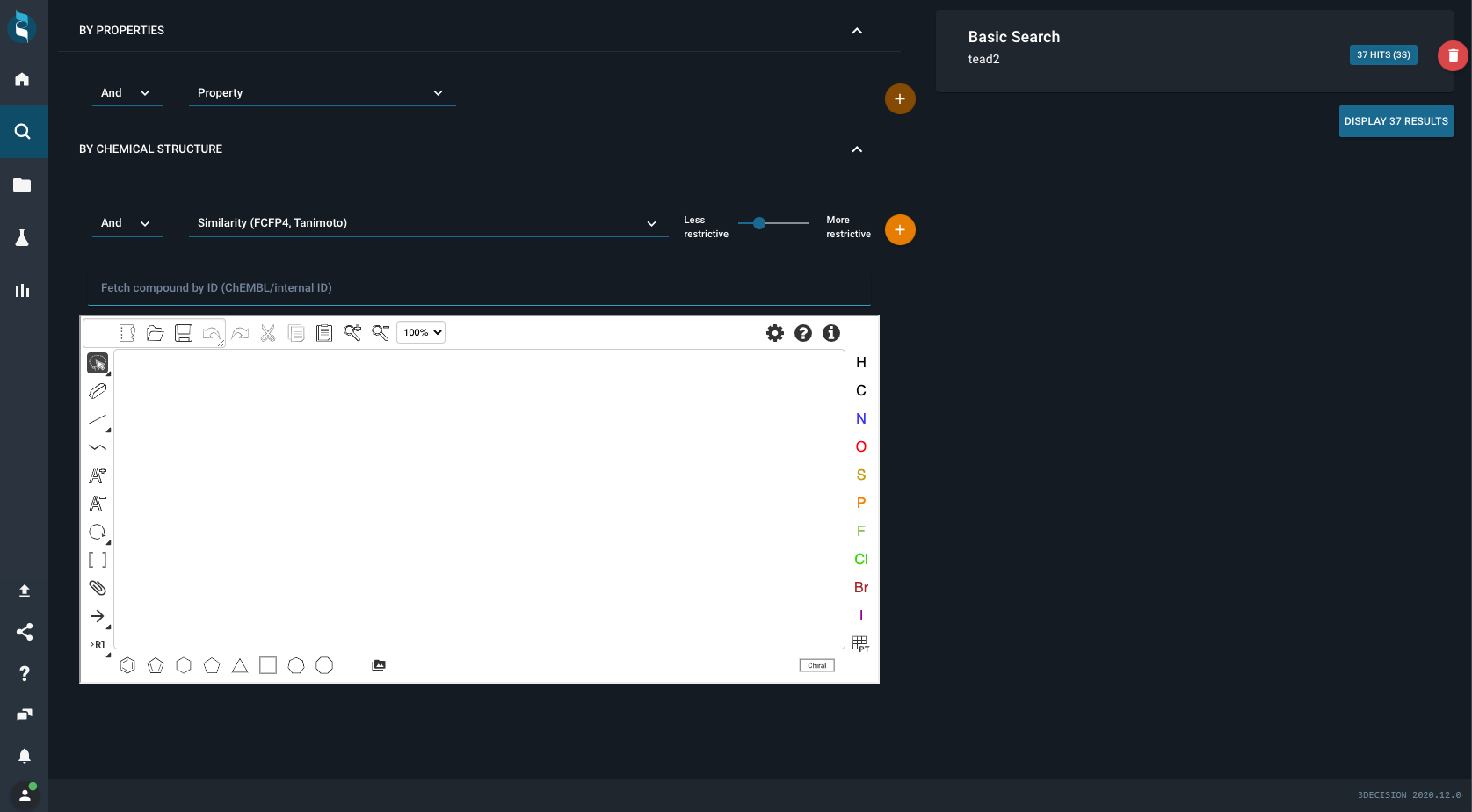
On the right side, you might see (or not) an entry saying Basic Search. This happens only if you already did a simple text search before opening the advanced search.
The advanced search page is split up in two main parts:
- left part: query building space - here you can define all queries you would like to use
- right part: query evaluation space - here you can get an overview of the results retrieved from all of your subqueries
Pre-created queries ban be deleted if needed by clicking on the red wastebin next to the query.
¶ Defining a text/numeric subquery
If you want to search by text or numeric data, annotations, space groups, resolution, protein families etc you can set up a property query on the left topmost side:
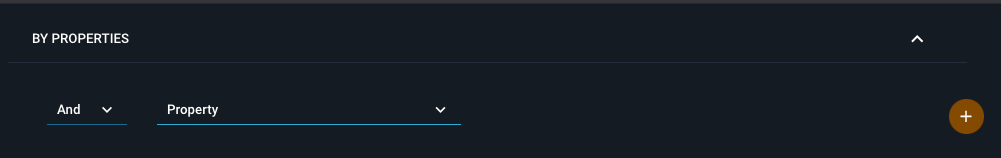
Simply open up the dropdown menu to select which property you want to search for.
Available properties are divided in five sections:
STRUCTURE
- Source
- Title contains
- Structure ID
- Project
TARGET
- Protein Name
- Structure Code
- Uniprot
- Gene Name
- Protein Family
LIGAND
- Residue Code (three-letter code)
- SMILES
- SMILES and similars
- Ligand ID (internal identifier)
PROPERTY
- Experimental Method
- Resolution (smaller, bigger or equal)
- Space Group
ANNOTATION
- Any Annotation (any of the following annotations)
- Structure Entry Annotation (of the PDB structure itself)
- Structural Annotations (annotations on some residues for structural description (ex: DFGin, DFGout) not available yet)
- Sequence Annotations (annotations on sequence residues, on the level of the biomolecule, not specific to one structure, could be preassigned or user defined sequence annotations, all prosite annotations are in this category too (ex: DFG position))
- Ligand Annotations (can be internal ID)
If you define for instance a protein family query, you can subsequently select which protein family you want to search for from ChEMBL target families. Once your query is set up click on the orange + icon on the right to add the subquery for evaluation to the right hand side.
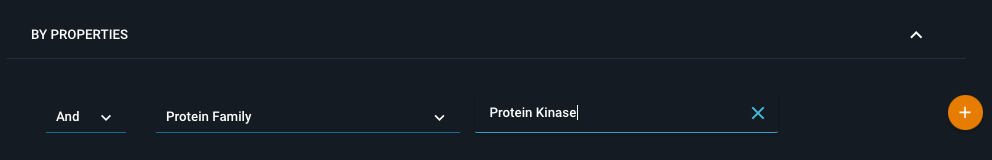
¶ Defining a chemistry subquery
Below the property query builder, you have also the possibility to create a chemistry-based query. This allows you to finally combine chemistry and text/numerical queries to achieve more powerful search queries.
To define a chemistry query you need to sketch or import a molecule or a fragment.
Next you can either modify the molecule or search for that molecule by using the features available in the molecular sketcher.
In this example, search for structures containing a acridine as a substructure. Sketch the acridine moiety and then open the dropdown menu above the sketcher stating Select a method- and select Substructure.
When clicking on the orange + icon the query will be accepted as subquery and get evaluated.
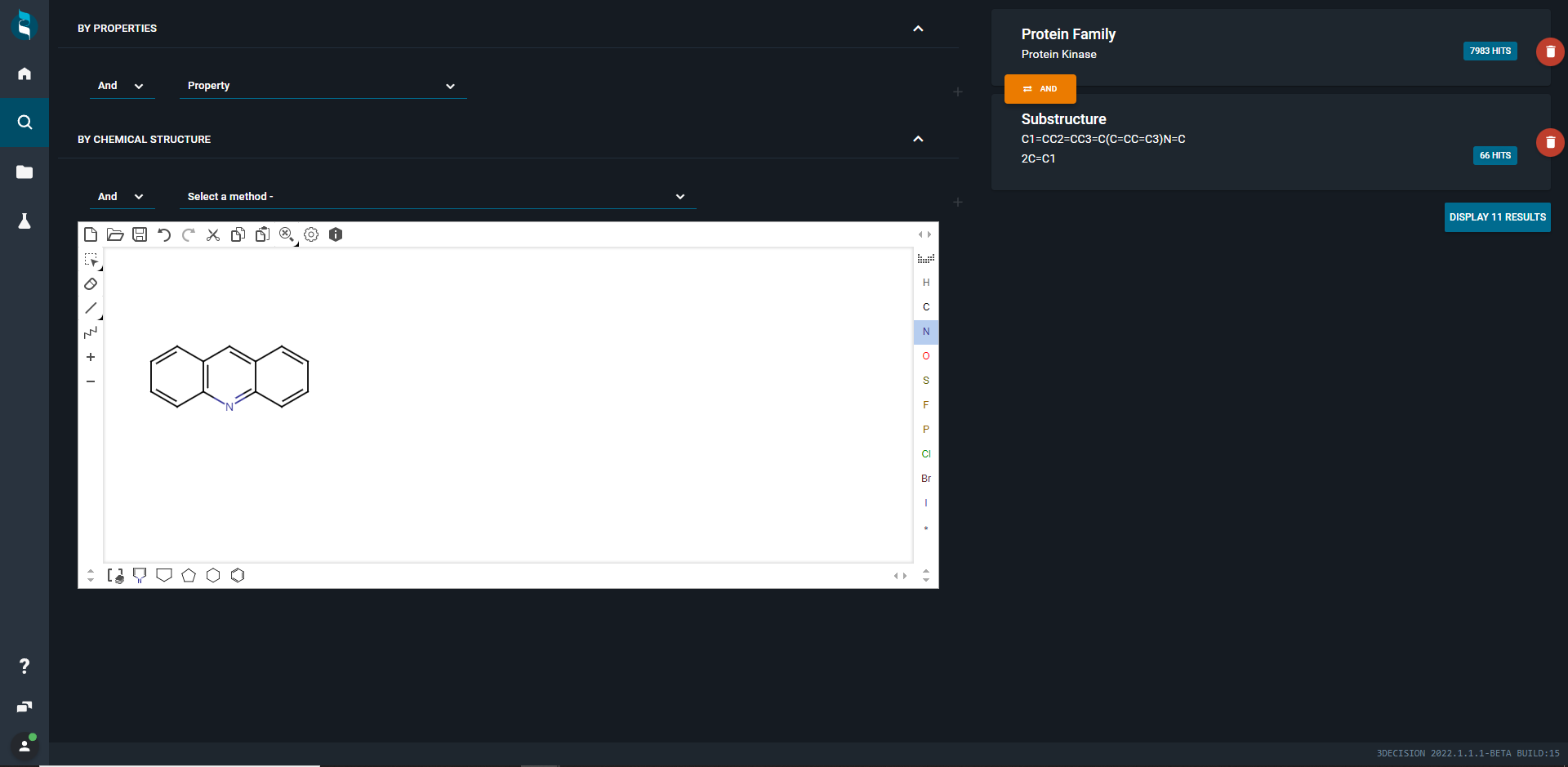
The available search options for chemistry searches are:
- Exact : search exactly for the same molecule
- Exact(+Isomers): search for the same molecule and its isomers
- Exact(+Tautomers): search for the same molecule and its tautomers
- Exact(+Tautomers & Isomers): search for the same molecule, its tautomers and its isomers
- Similarity: search for similar molecules using ChemAxon fingerprint and Tanimoto (full molecule comparison) [default threshold is 0.65]
- Substructure: search for molecules containing the exact substructure
¶ Evaluation of a subquery
Every time you add a new query to the query evaluation, it gets evaluated immediately.
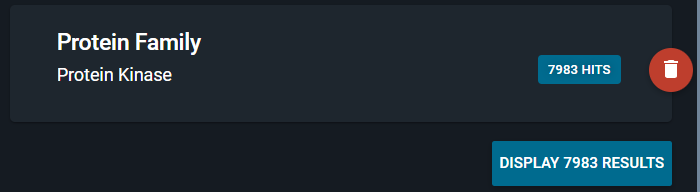
For the protein family search, we see immediately how many structures are retrieved with that search, in this example 7983 hits.
Adding a second subquery, will also evaluate this subquery independently. Here all structures with a resolution better than 1.5A are searched for, 18494 hits are found.
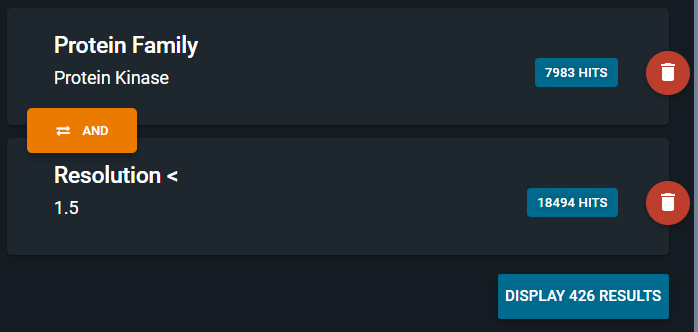
¶ Combining subqueries
If you use several subqueries like in the example above, they are by default combined with a logical AND operator. Here we will search for structures containing a protein that is in the protein kinase family and that has a resolution better than 1.5 Angströms. In total, we will retrieve 426 structures from that search.
You can also use AND NOT operator between subqueries.
Simply click on the Display Results button to show all results in the standard 3decision search results page.