¶ Introduction
The Pocket Similarity Search in 3decision enables users to search for similar pockets or similar sub-regions of the binding site in the entire 3decision knowledge base (proprietary and public structures, including AlphaFold models).
This feature can be used to collect all structures with similar protein conformations, to find potential off-targets, or to find a new chemical matter for drug design idea generation.
¶ Find and select structures
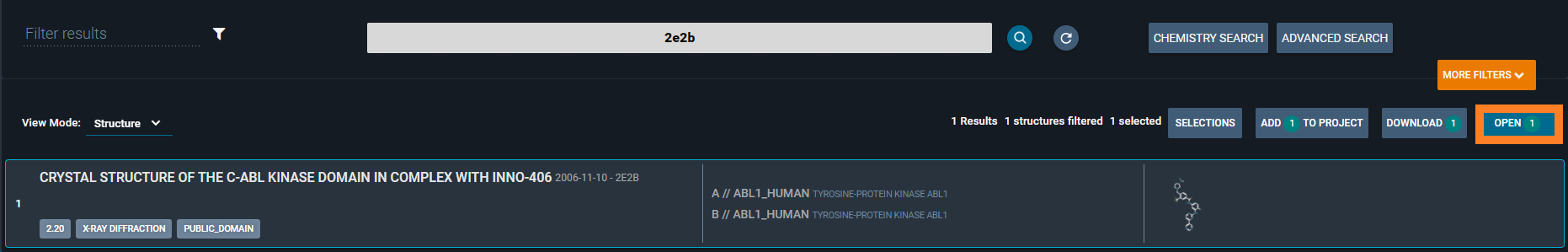
Find the structure using the search bar on the Homepage and click on the lens-icon to run the search. Select a structure of interest and press “Open” on the top right corner to enter the Workspace. For more details, check Search page
Type 2e2b and click on OPEN.

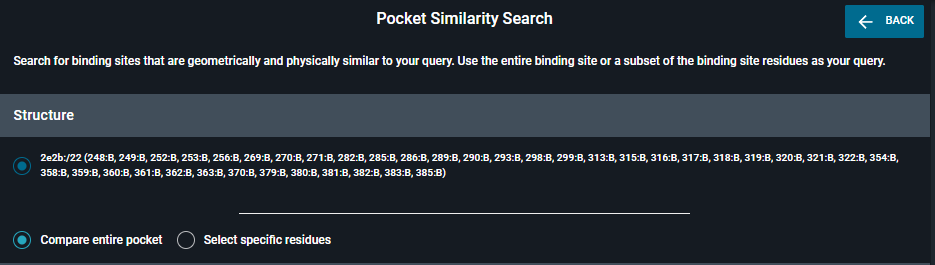
With the new Pocket Similarity Search engine, two options are proposed. You can do an entire pocket or a subpocket search.
¶ Compare entire pocket
You can go at the top in the navigation bar or click on Pocket Browser in the dropdown menu, and select the pocket of interest in the table. All the residues that are lining the pocket will be considered for the search. The list of ResidueNumber:ChainName is given at the top of the panel.
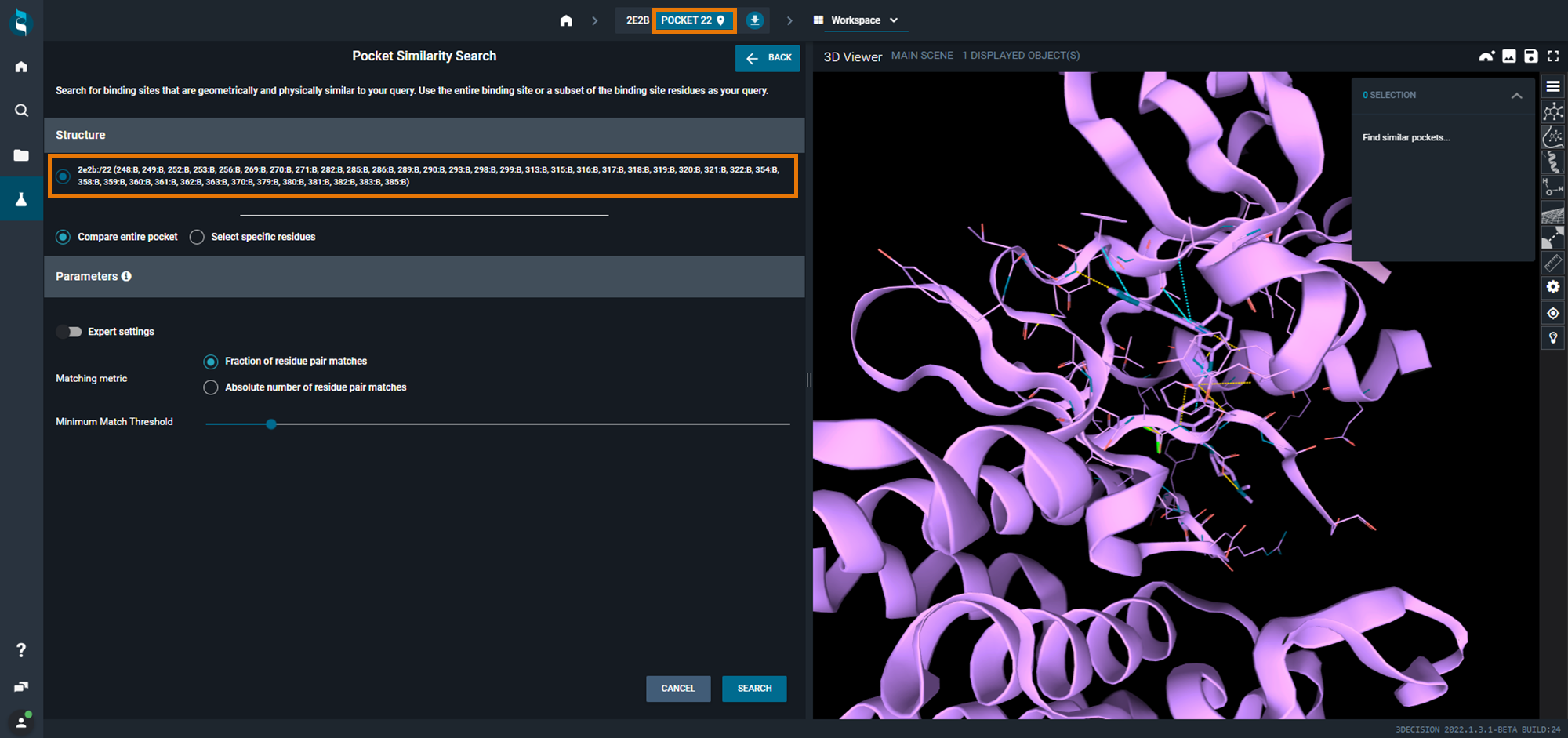
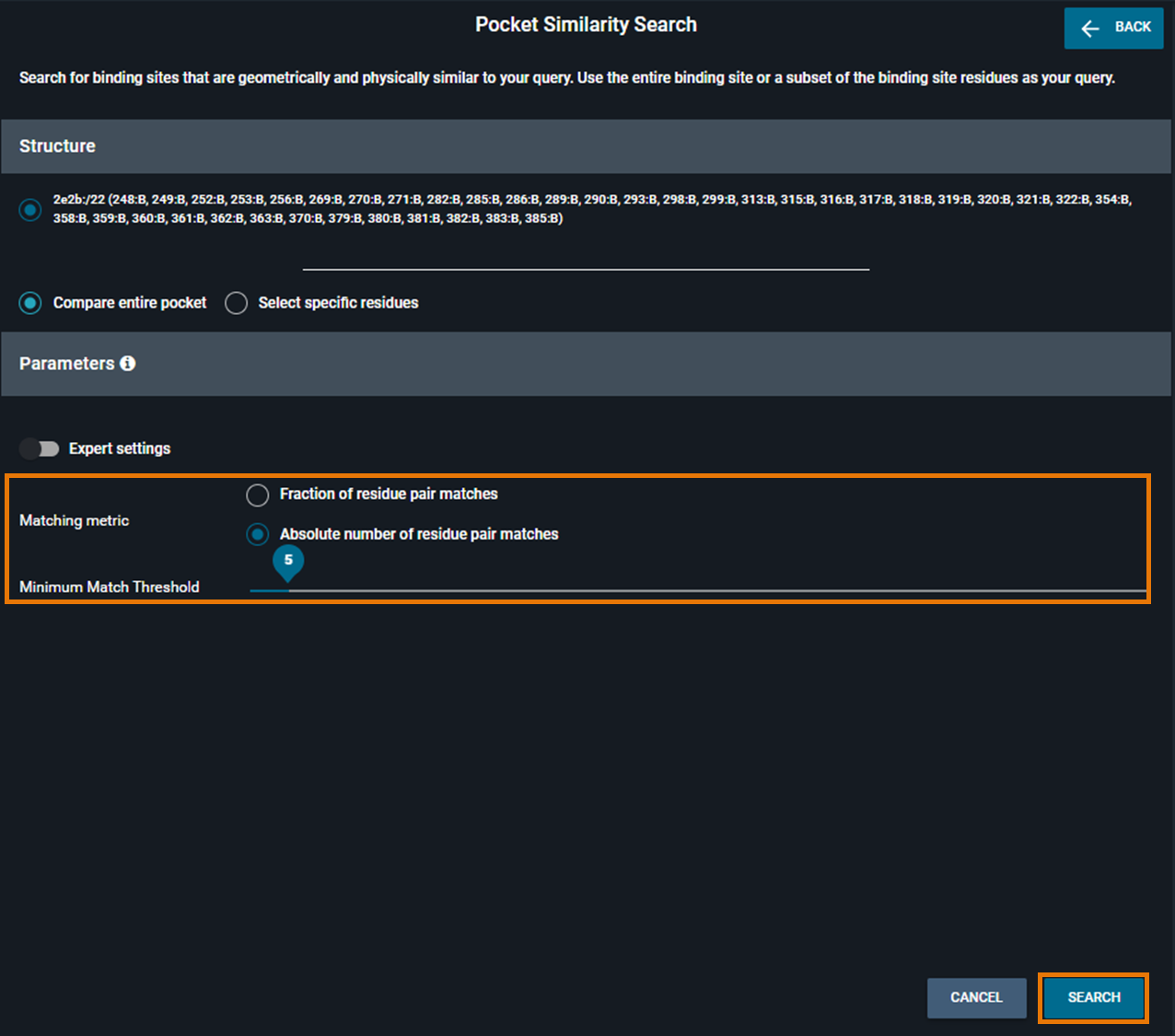
Refine the query and start the search
You can modify the default parameters. You can either use Fraction or Absolute number as Matching metric. Then, you can change the value on the slide bar from 0,1 to 1 for Fraction and from 3 to 50 for Absolute.
For this example, we used 5 as an absolute number of residue pair matches. Then, click on SEARCH.
¶ Select specific residues
To define the subpocket of the chosen structure, select amino acid residues directly on the 3D Viewer in the following ways:
- Select amino acids one-by-one with the left mouse click
- Use Alt+Left mouse click to select all the residues within 4.5 Å of a desired ligand atom
Selected amino acids are presented in white.
The total number of selected amino acids is displayed on the action menu on the top right corner of the 3D viewer (SELECTIONS). To refine your list of residues prior to the search, click on the little arrow near “SELECTIONS” and go to “Find similar pockets”.
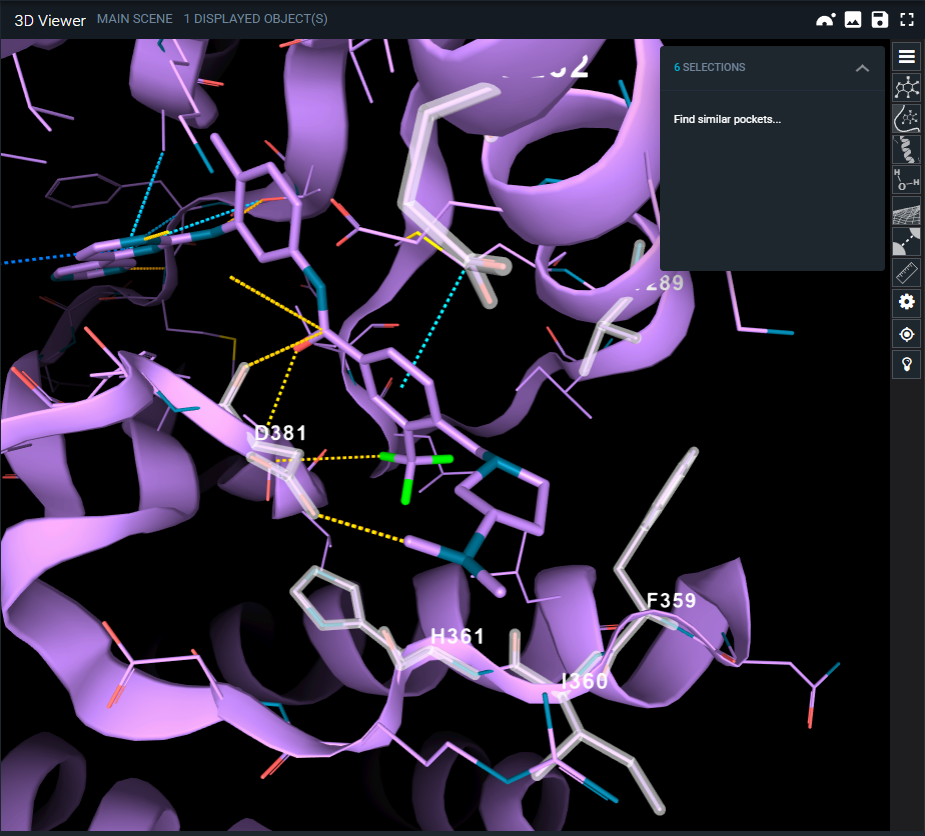
Only residues that are part of the selected pocket could be considered during the search.
Refine the query and start the search
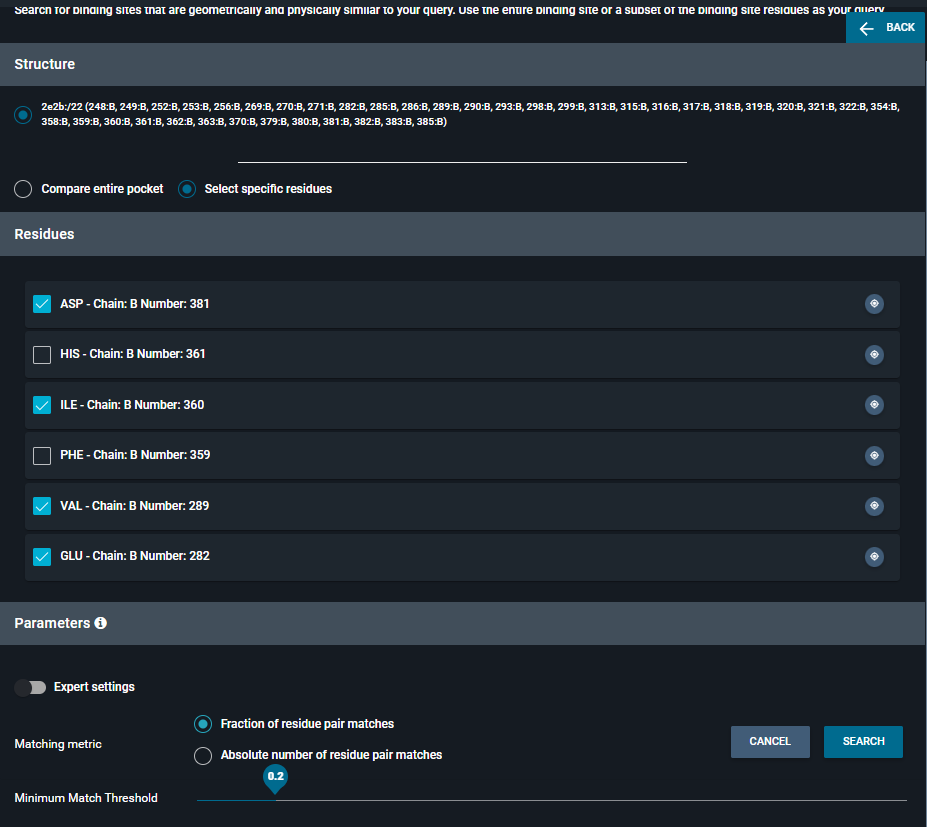
The subpocket search panel will appear on the left side of the screen. In the “Residues” section, deselect amino acids you want to exclude from your search. Don't worry - at this point, it is still possible to add amino acids from the 3D viewer (as explained above).
You can either use Fraction or Absolute number as Matching metric. Then, you can change the value on the slide bar from 0,1 to 1 for Fraction and from 3 to 50 for Absolute number.
For this example, we used 0,2 as an fraction of residue pair matches. Then, click on SEARCH.
¶ Get the results
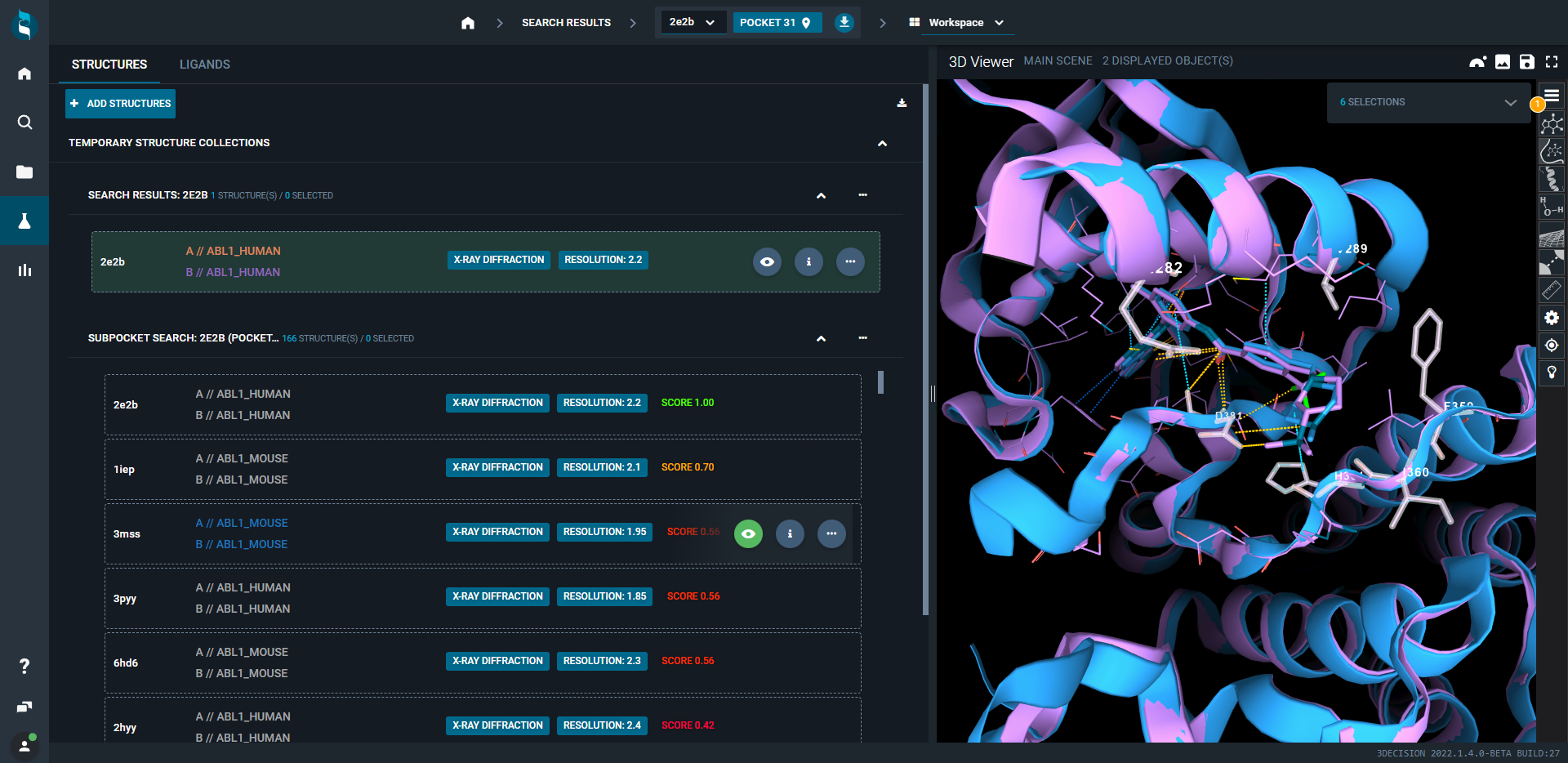
For each structure, a match score is assigned (maximum 1.0 for a perfect match).
¶ Structure view mode
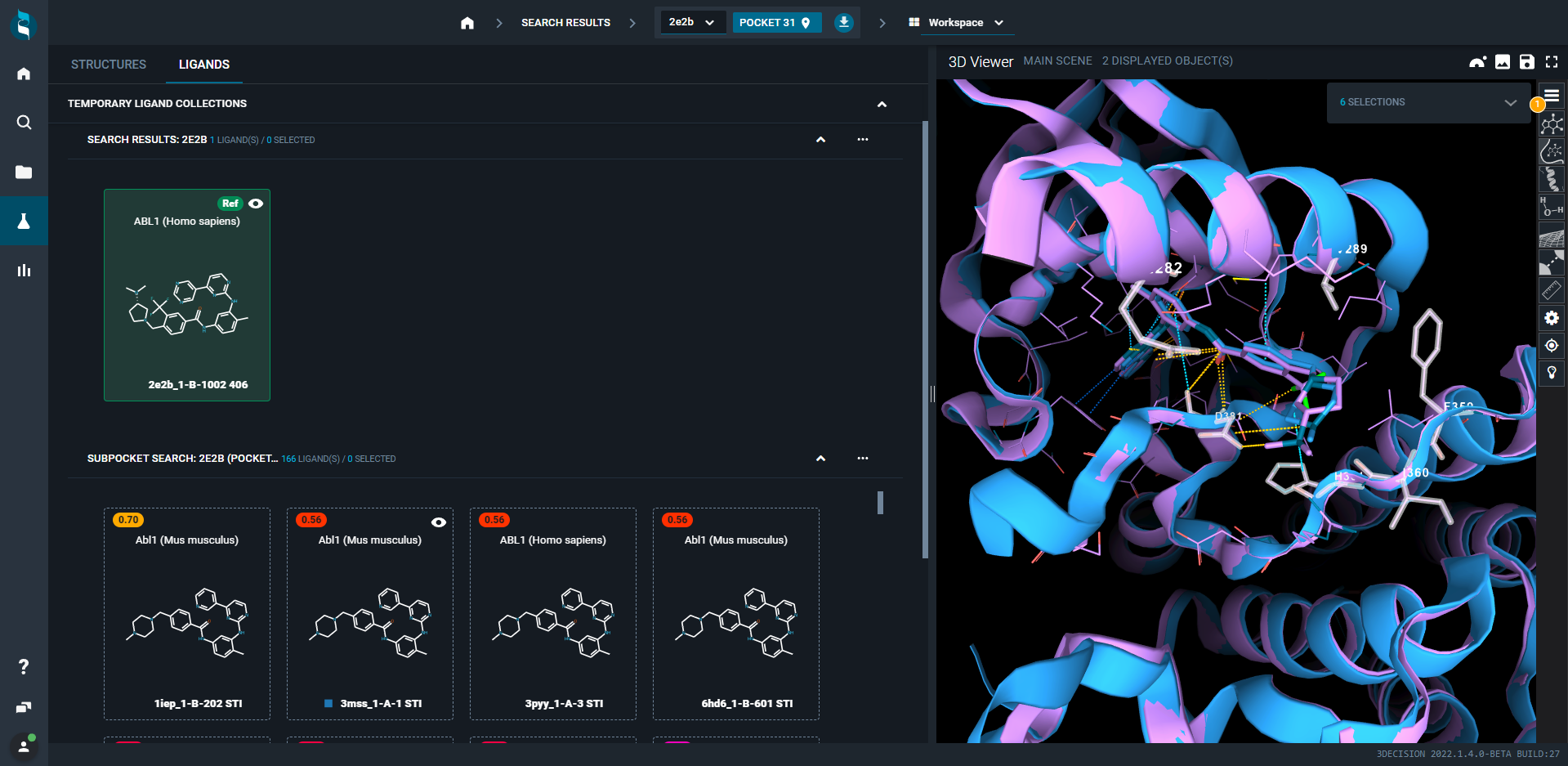
The results are presented on the left panel in a new collection in the Workspace - Structure tab. The new structure collection is sorted by the calculated score by default. To compare the search results with the query, superpose structures of interest by clicking on the eye–icon.
¶ Ligand view mode
It is also possible to browse the results from the LIGANDS Tab by clicking on the top left of the screen and to superpose structures in the same way (eye-icon).