¶ Introduction: Superposition methods
In 3decision, you can easily overlay and compare protein structures. We support two superposition methods, to provide flexibility for structure alignment and allowing you to choose the most appropriate option for your specific use case:
- Superposition by pocket, more suited for comparing small molecules binding sites of similar proteins;
- Superposition by chain or residue selection, adapted for the comparison of protein complexes and antibodies.
You can choose between both superposition methods in the Workspace and when setting up Projects, as detailed later in this page of the documentation. The main difference between the two options:
- superposition in the Workspace is performed on-the-fly, and it is not saved in the database, so when you close the application, you will loose the alignment.
- superposition within 3decision Projects allows you to save the alignment in the database, and easily share the pre-aligned structures with your colleagues.
¶ Superposition by pocket
This superposition method is based on the pockets that are automatically detected on each structure during the structure import process (see this page for more details).
The superposition by pocket uses the reference pocket of the reference structure as input for the sequence alignment. When launching the superposition:
-
If the two structures contain the same identical protein, the binding site residues are selected on both structures, and a distance-weighted optimization (reference link) is run to superpose the two structures. This superposition algorithm allows to account less for very flexible parts of the structure and focus on the more stable parts.
-
If the two structures contain different proteins, the first step is to identify the best matching pocket. To do that, a sequence alignment of all biomolecules composing the reference pocket versus all biomolecules in the structure to superpose is run. The best matching alignment is kept. Then on this identified biomolecule, the pocket overlapping the most with the pocket on the reference structure is selected. From this, the residue mapping allowing the distance-weighteded superposition is extracted.
The superposition by pocket is successful when there is enough overlap between the two pockets to overlay, and when the arrangement in space is similar. Therefore, it is particularly adapted for the comparison of binding sites of similar proteins.
¶ Superposition by chain
This superposition method exploits the structure-to-sequence mapping that is automatically performed during the structure import process. The sequence mapping analysis maps each structure chain to one or several biomolecules (i.e. UniProt entries or generic sequence identifiers).
The superposition by chain is based on a sequence alignment between two structure chains provided by the user. The structure sequences are either entire structure chains (mapped to a specific Uniprot code or generic sequence identifier) or a selection of residues on each structure (e.g. a residue range).
The superposition is optimal when the two chains correspond to the same protein but has a certain degree of flexibility to allow for the overlay of close ortholog sequences. If the two chains correspond to different proteins, a sequence alignment is performed and the residue range (if provided) is extracted from the alignment in order to drive the superpositioning.
This superposition method is particularly suited for the comparison of proteins with multiple chains.
¶ Superposition in the Workspace
You can easily assess structural differences between two or more structures in 3decision by overlaying them in the 3D Viewer.
From the Workspace, you can superpose structures on-the-fly, in a few clicks, with the possibility of selecting the superposition option that is more suited for your analysis.
The superposition within the Workspace is extremely intuitive and fast to set up, so it allows to quickly analyze and compare structures. However, this superposition it is not saved in the database, so when you close the work session, you will not be able to restart working on the overlayed structures. If you want to save the alignment, and be able to share it with your colleagues, you should work in the context of Projects (as described in the next section of this page).
¶ Superposition by pocket
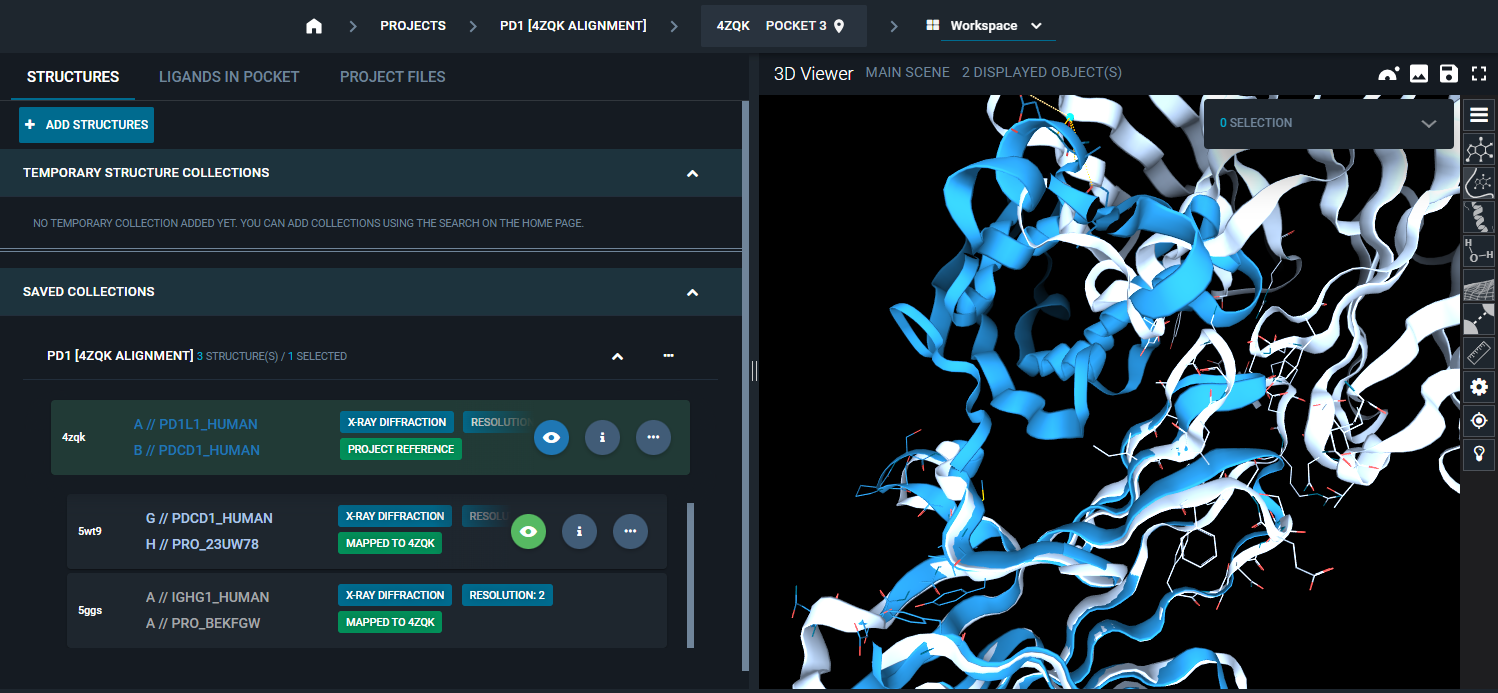
For superposition by pocket to be successful, it is important that you set the reference pocket you are interested in for the alignment. You can see the default reference pocket of your reference structure at the top of the page.
If you want to change it, you can do that from the Pocket Browser.
Once set the reference pocket for the alignment, you have two options for superposing structures by pockets in the workspace:
¶ 1. From the "Eye" icon
Hovering with your mouse on the structure card of the structure you want to overlay, you have the "eye" icon appearing. Clicking that button, the superposition by pocket will be performed.
![]()
This is because the superposition by pocket is the default superposition method in 3decision, and so it is the one launched automatically from the "Superposition" button of the workspace.
¶ 2. From the Structure Actions menu
On the structure card of the structure you want to overlay, you can select the "three-dots" icon (structure actions).
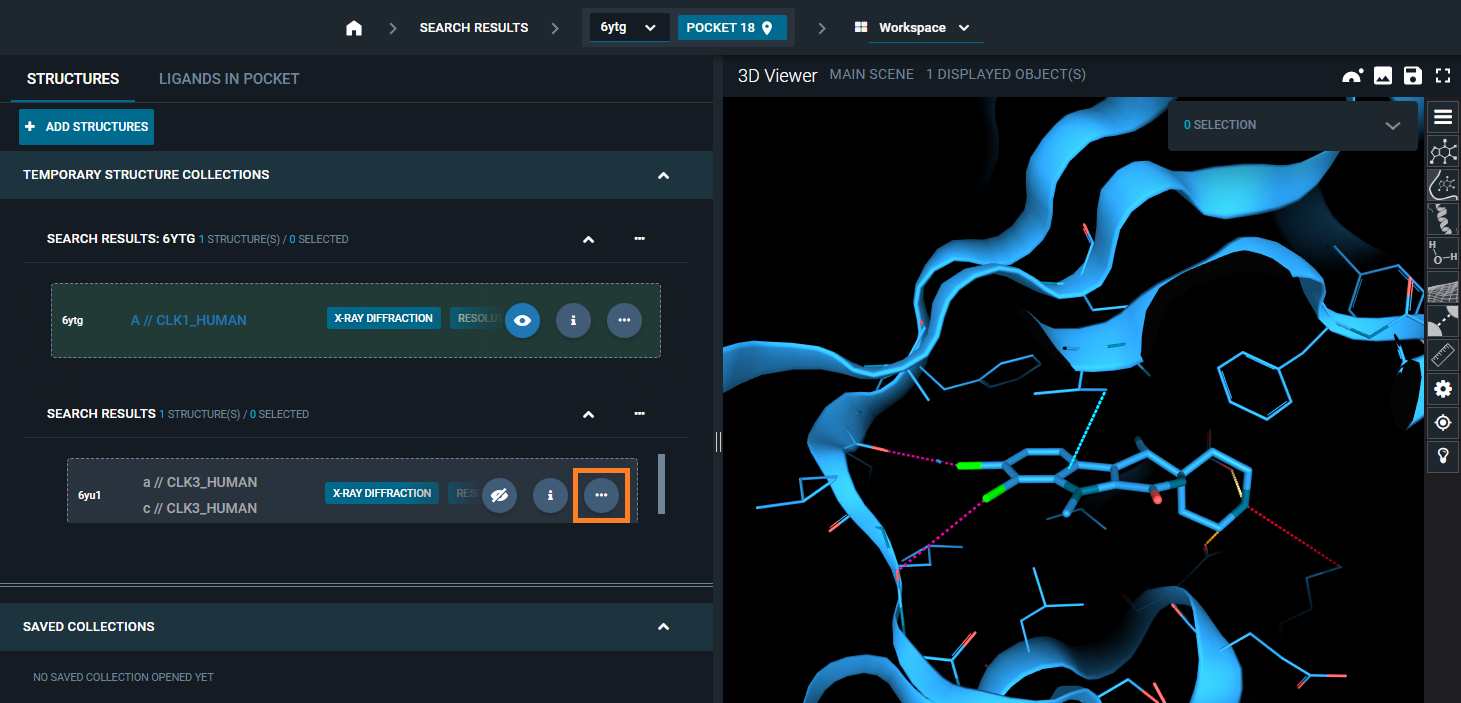
From the structure action menu that appears, you have a the option: Superpose. Selecting this, you can choose between two superposition options. By selecting Superpose locally (Automatic), superposition by pocket using the reference pocket for the alignment will be performed.
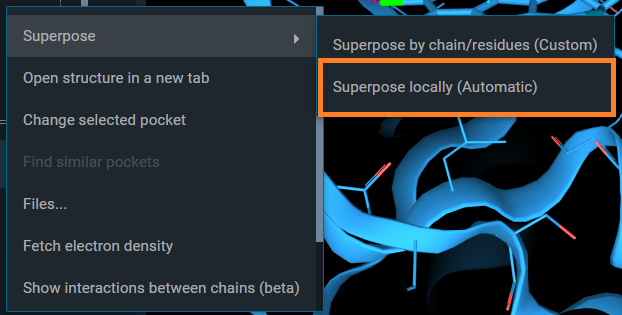
Both ways, you end up with the same aligment of structures.
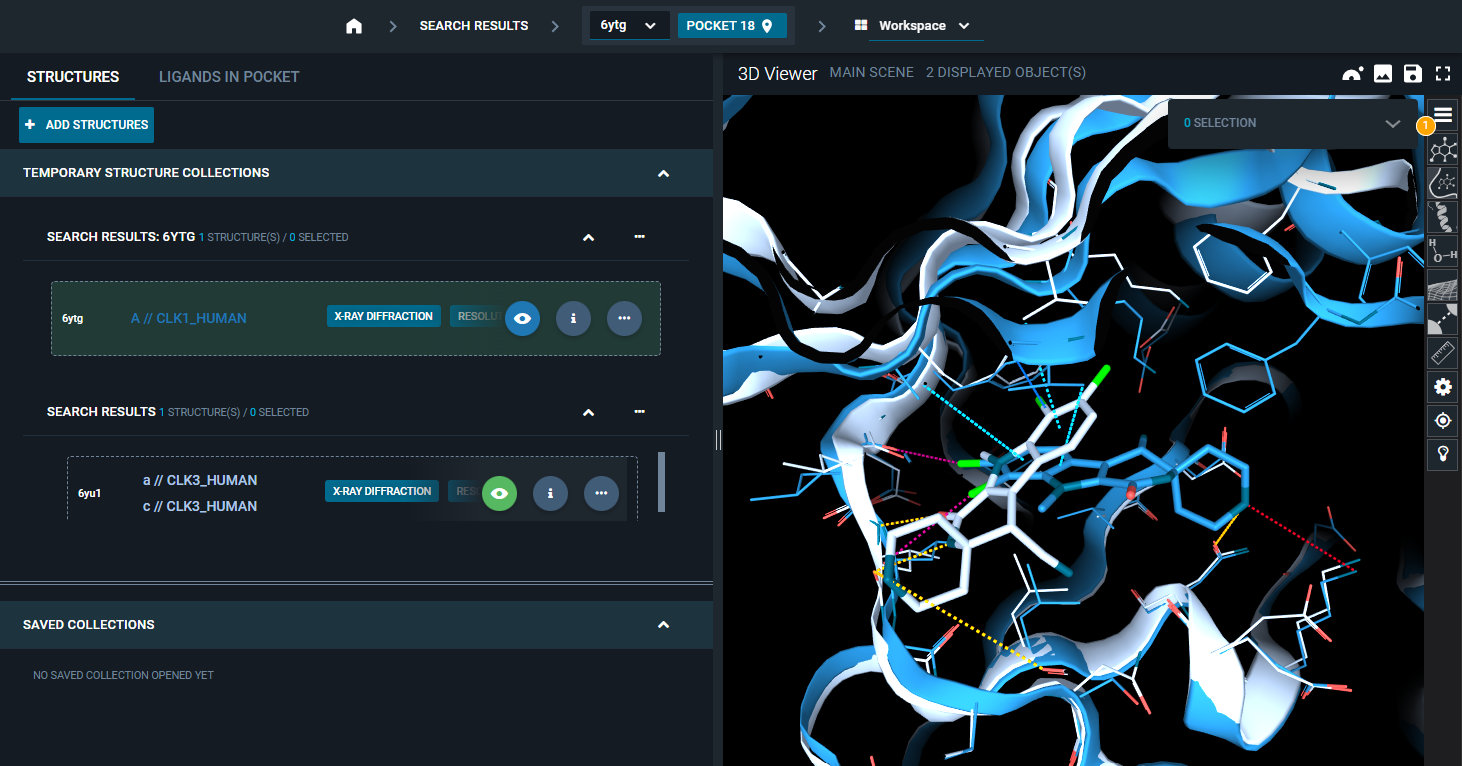
¶ Failure of superposition by pocket
If the superposition by pocket is not successful (e.g., pockets too different), you will get a pop-up with an error message and other action options:
Manually select: allows to run superposition by chain, manually selecting the protein chains (and residue ranges) to superpose.Open in new tab: it will open the second structure in a new 3decision tab.Show in 3DViewer not superposed: the second structure will be added to the 3D Viewer, but without any alignment to the reference structure.Nothing: the second structure will not be displayed.
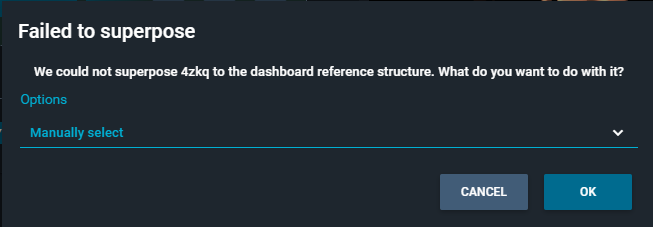
¶ Superposition by chain/residue selection
In alternative to superposition by pocket, you can decide to perform the alignment based on a certain protein chain, or a specific region/domain of the protein.
To launch this superposition, you need to hover with your mouse on the structure card of the second structure, and select the "three-dots" icon (Structure Actions).
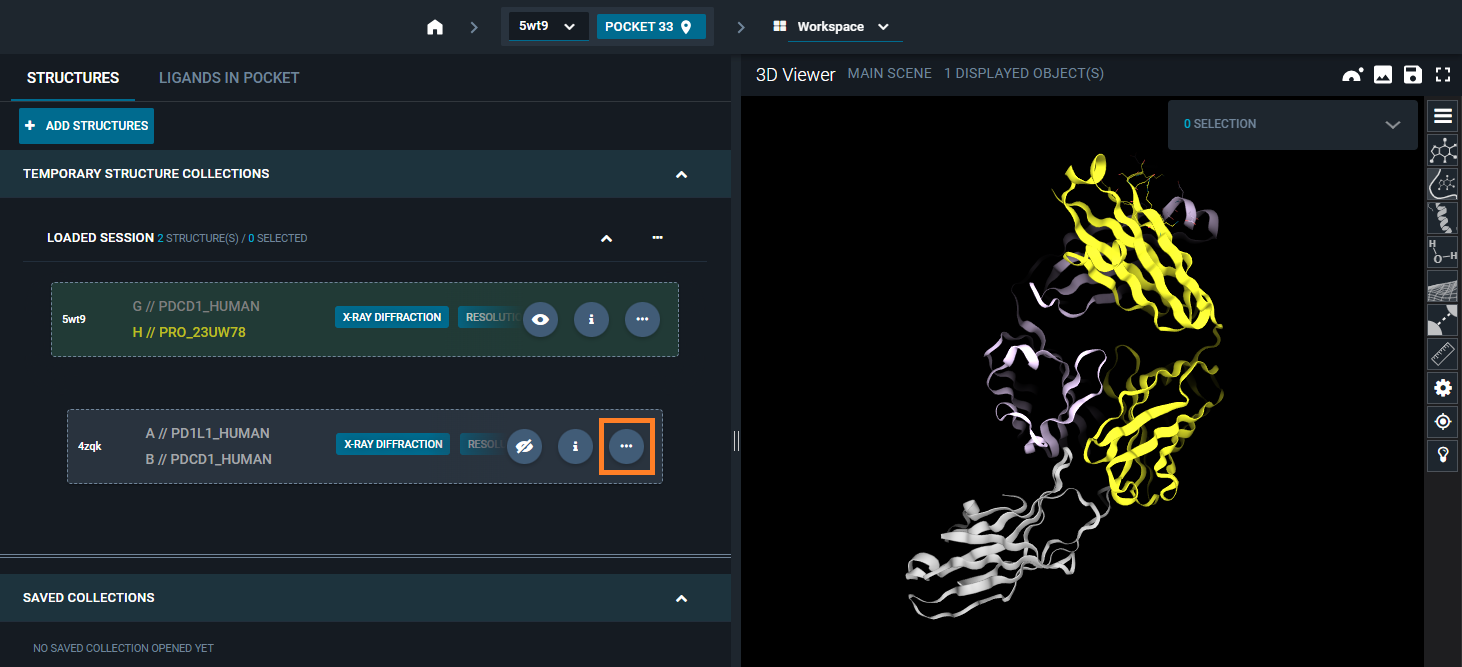
From the structure action menu that appears, you have the option: Superpose. From here, you can should choose Superpose by chain/residues (Custom).
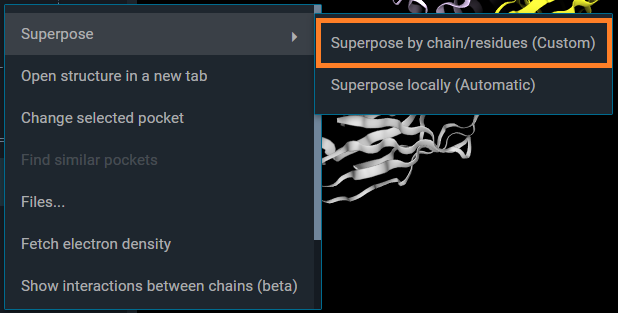
This opens a pop-up "Superpose by chain/residues", which allows to set the protein chain you want to use for the alignment for each structure. Optionally, you can specify a certain residue range, to perform the superposition on a specific region or domain of the proteins.
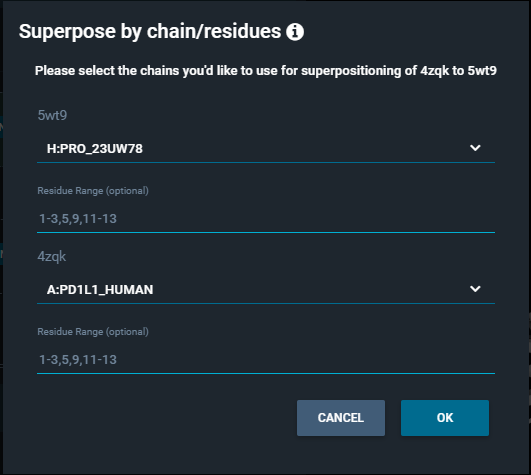
In this way, the superposition is not based on the reference pocket, but on the chains and residue selection that you gave as input.
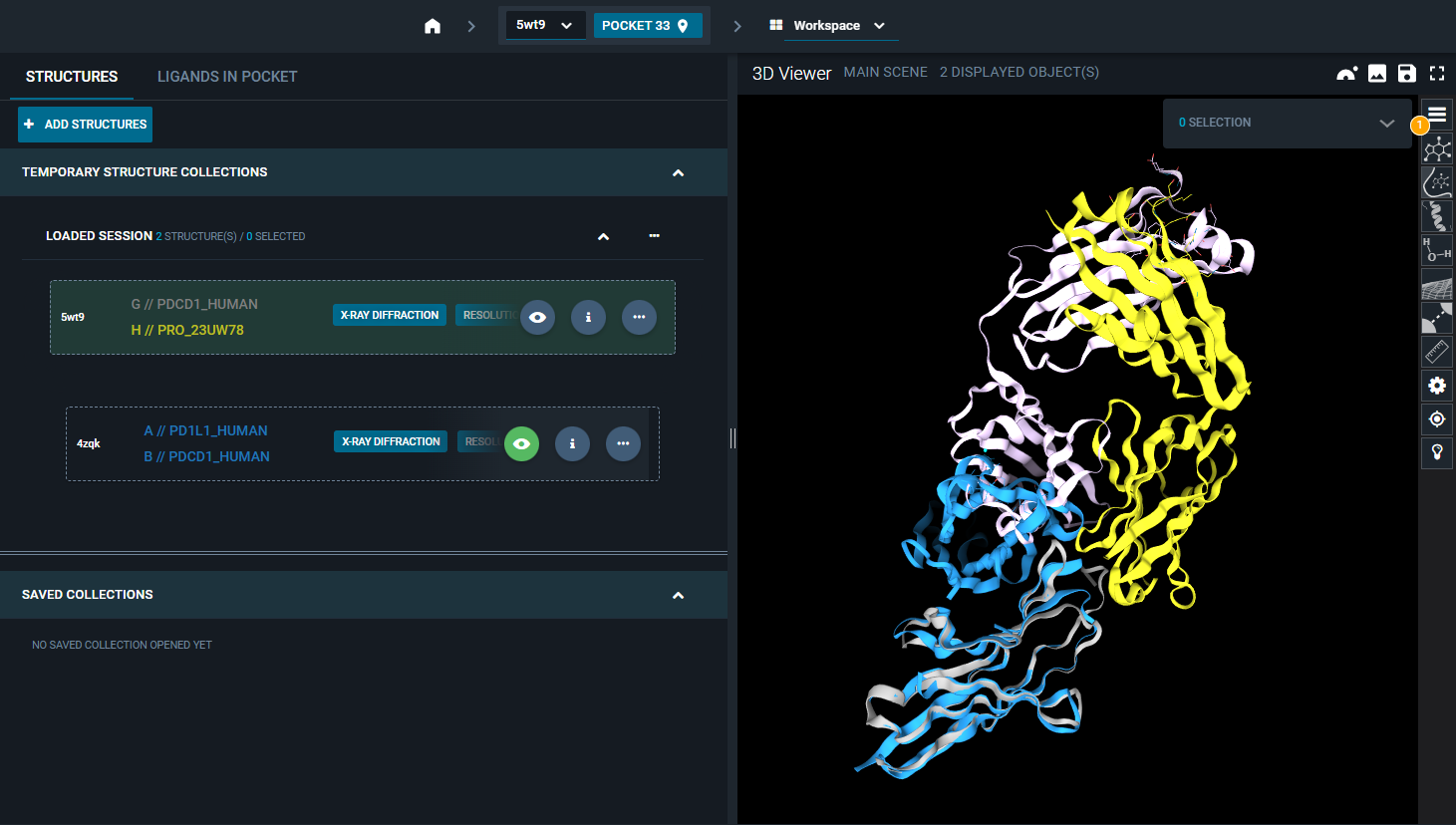
¶ Superposition in Projects
Within the 3decision Projects, you have the possibility to align a collection of structures saved in the project to a reference structure. This pre-alignement is saved in the database.
In this way, when you open the structures from the Project, these are already pre-aligned, and the superposition is not performed on-the-fly. Also, anytime you add a new structure to this project, it will be automatically pre-aligned to the set reference.
This is also an easy way to share overlayed structures with other colleagues within the drug discovery team.
More details on 3decision projects can be found here.
Once you are in the Project Overview, you can setup the project reference from the button on the top of the page.
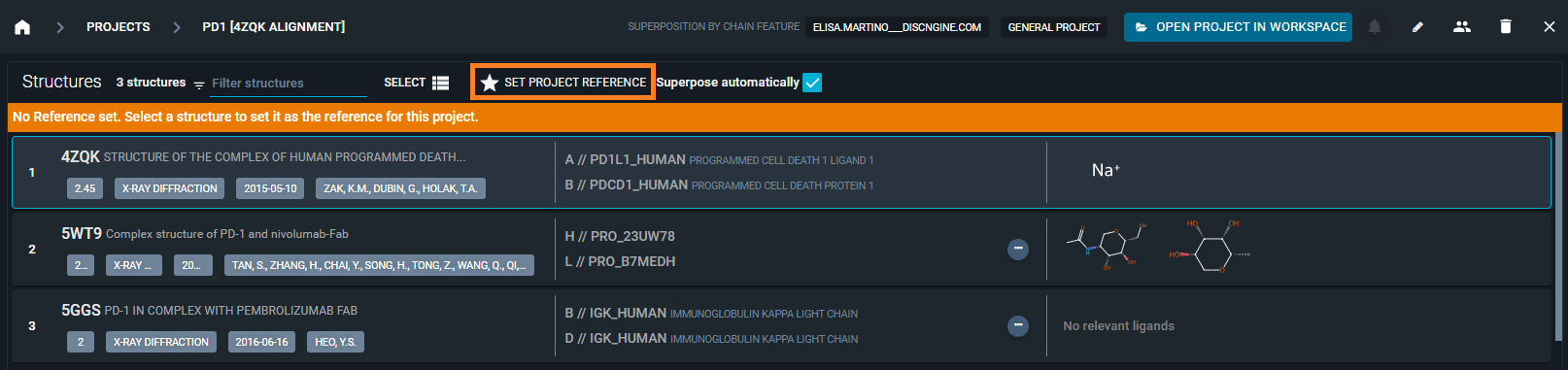
From here you have the possibility to choose between the superposition method to use for the alignment of the structures in the Project.
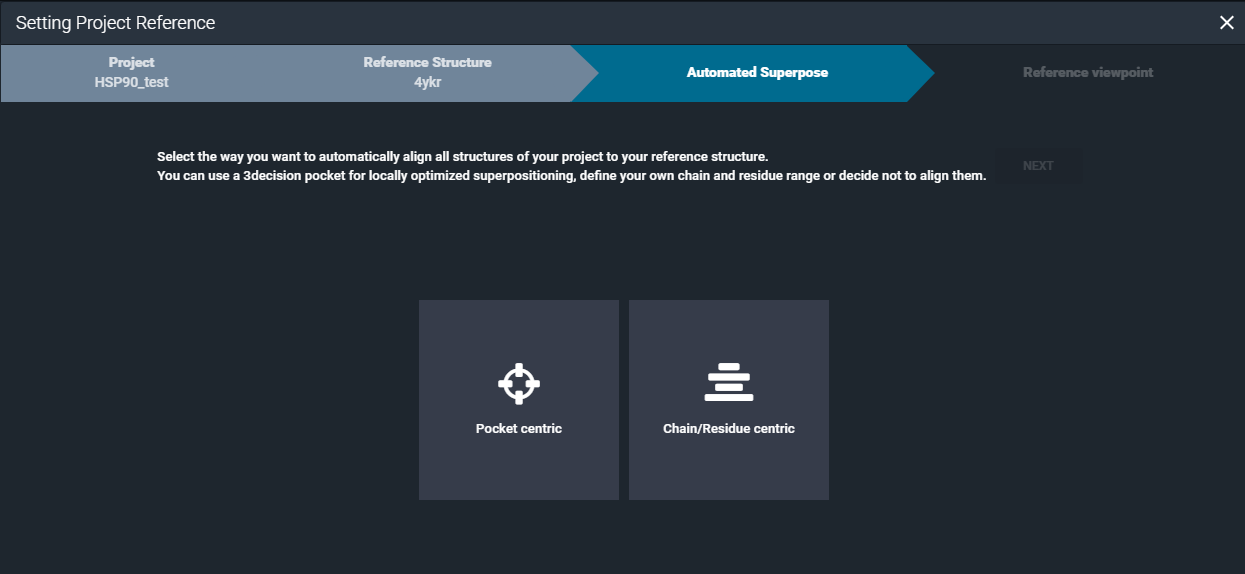
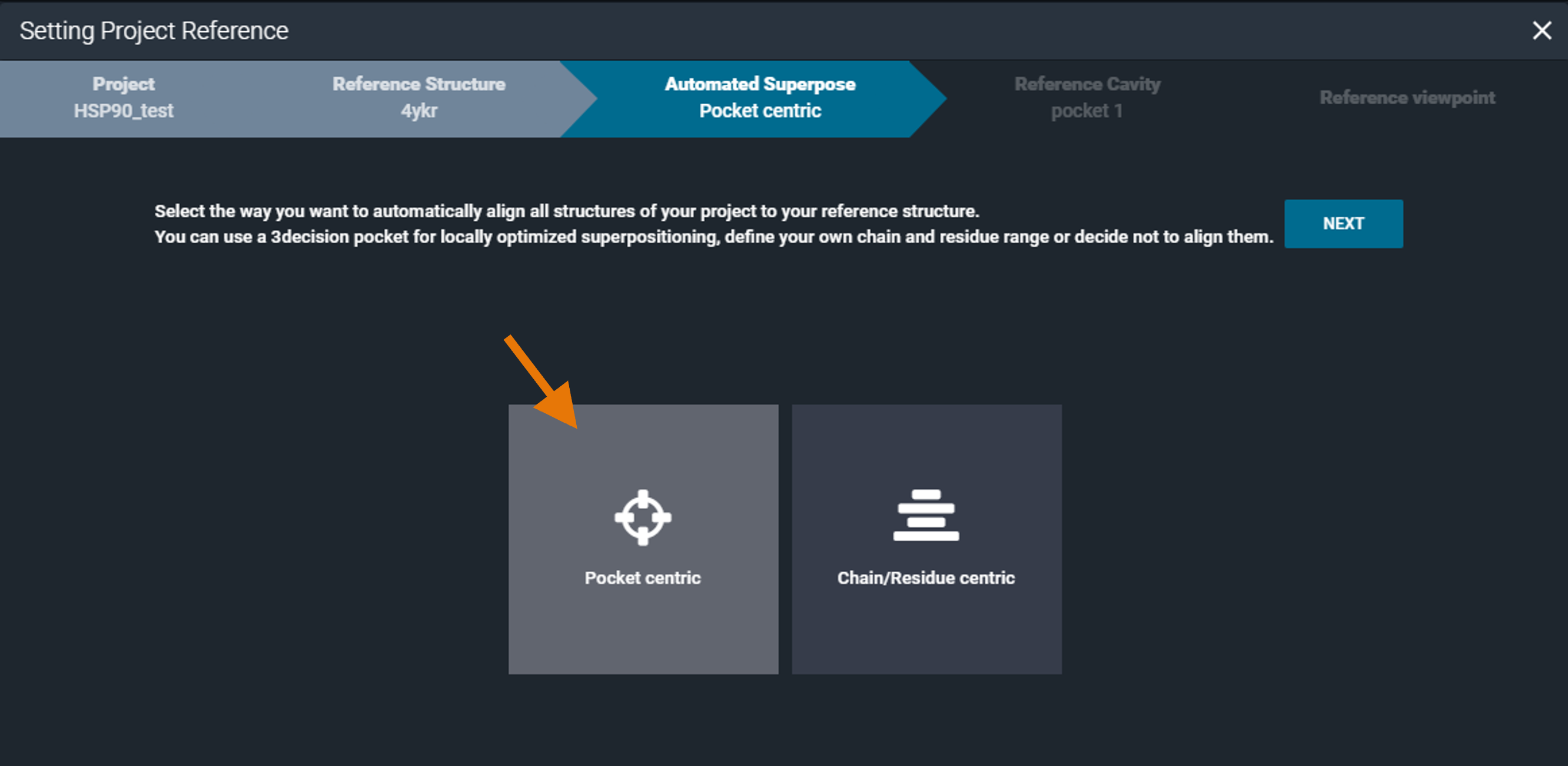
¶ Pocket centric superposition
By selecting the "Pocket centric" option, the superposition by pocket algorithm will be used for aligning the structures in the project.
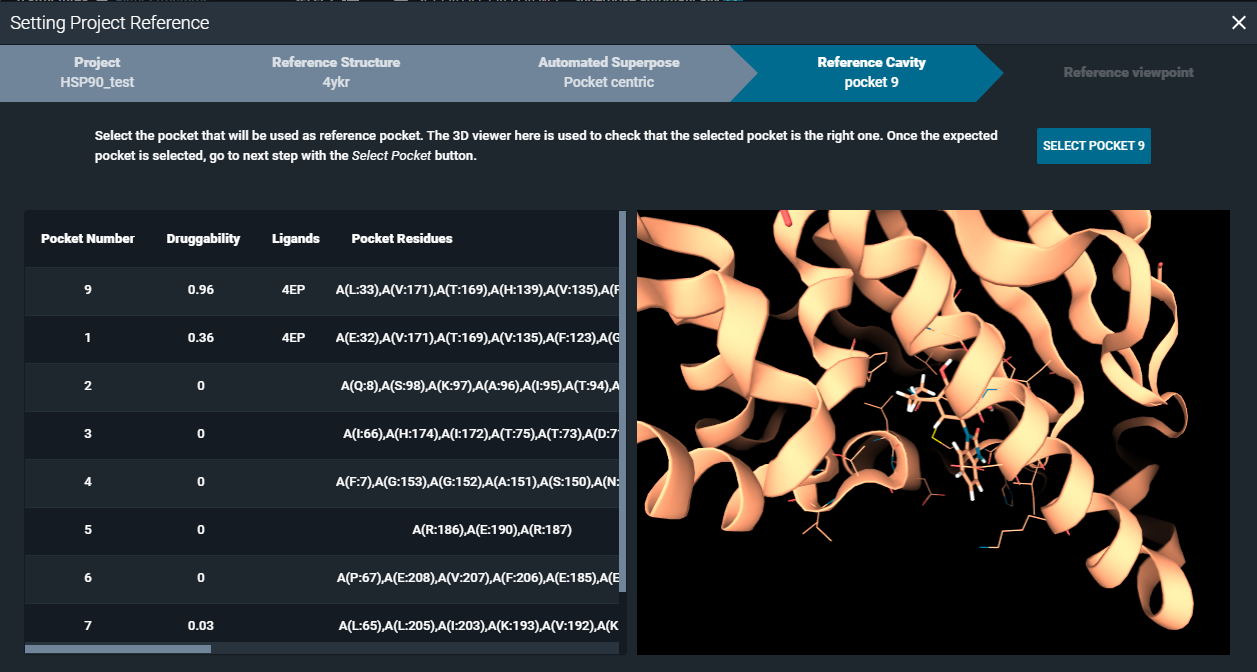
You need to select a reference pocket from the project reference structure. To help you selecting the most suitable pocket for the alignment, you have the pocket browser on the left side, and 3D Viewer on the right side of the window.
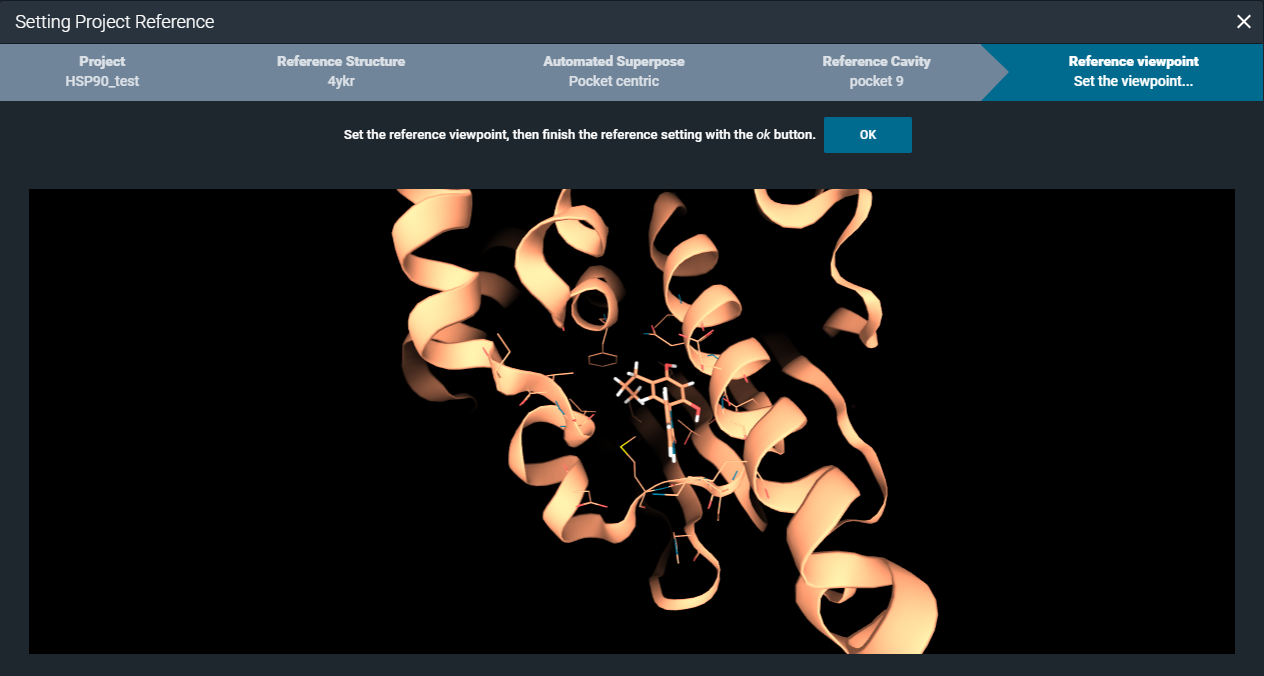
Finally, you can set a specific viewpoint, that will be used when opening the structures in the workspace from the project.
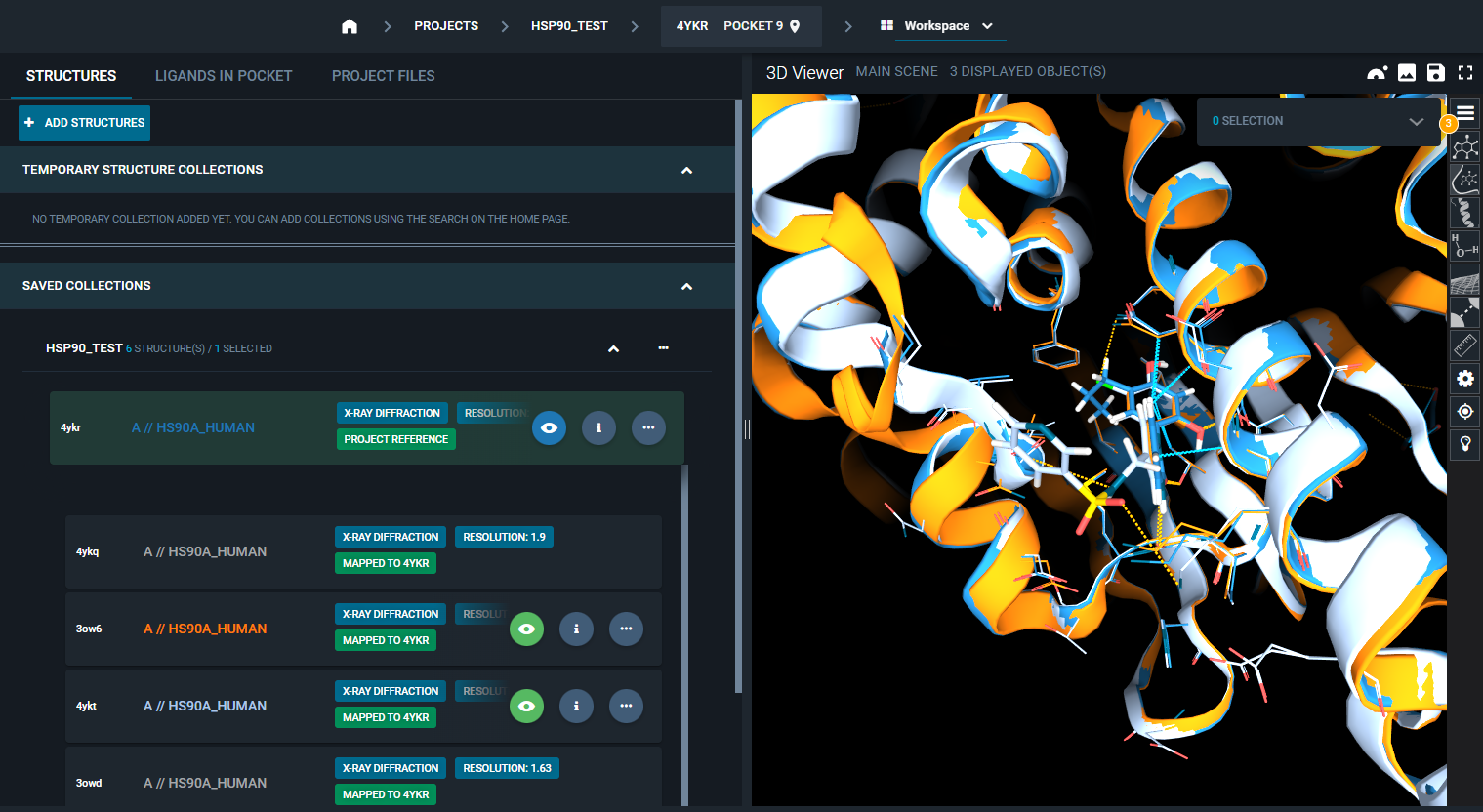
In this way, you have the collection of structures in the project (and any additional structures added to the project at a later time) pre-aligned to the reference pocket of the project reference structure.
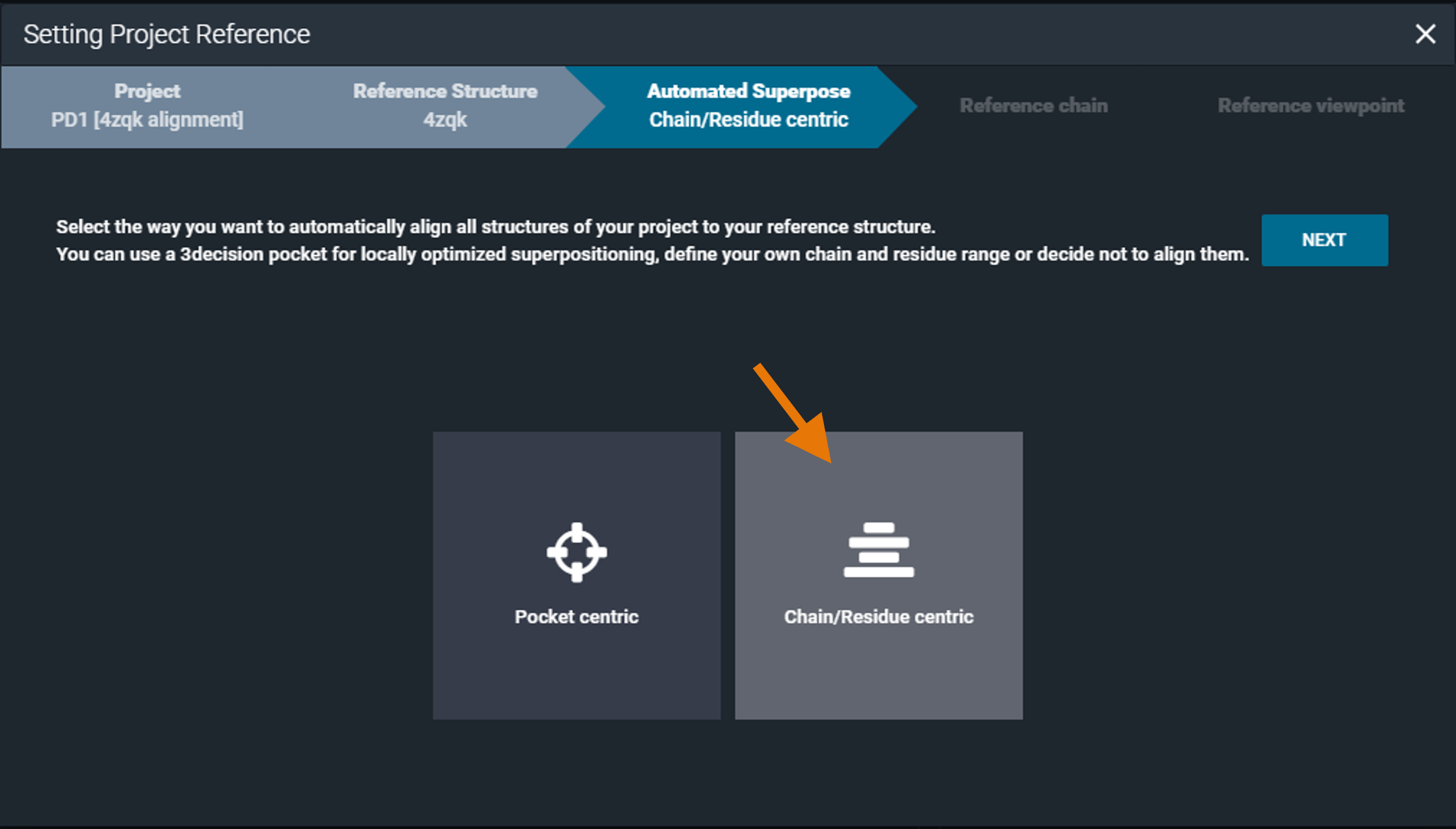
¶ Chain/Residue centric superposition
By selecting the "Chain/Residue centric" option, the superposition by chain algorithm is used for overlaying the structures contained in the project.
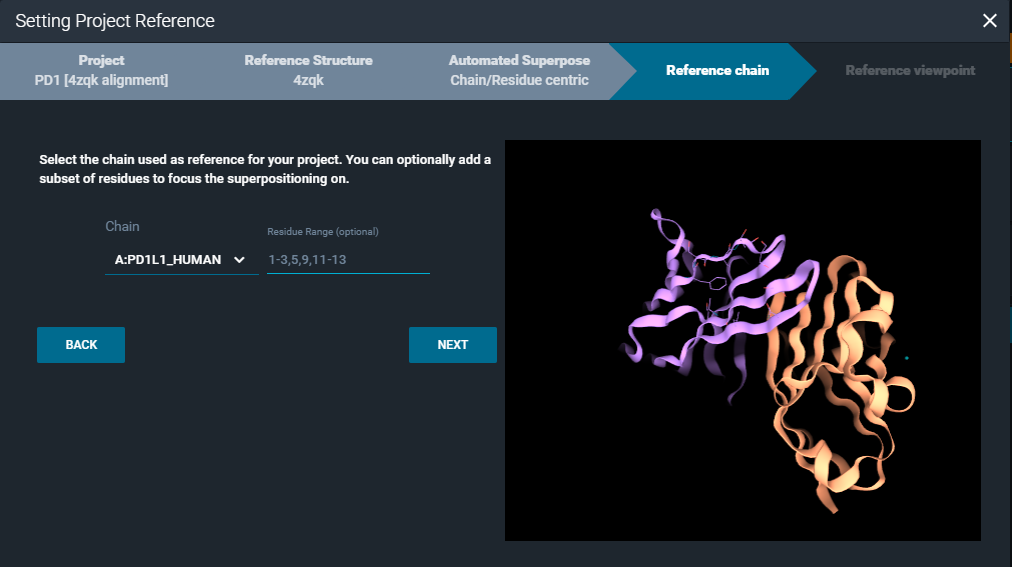
You need to select a reference protein chain from the project reference structure. Optionally, you can also define a specific residue range as a reference for the alignment. On the right side of the window, you have the 3D Viewer to support you in the selection of the desired protein chain/residues.
Finally, you can set a specific viewpoint for visualizing the project structures in the workspace.
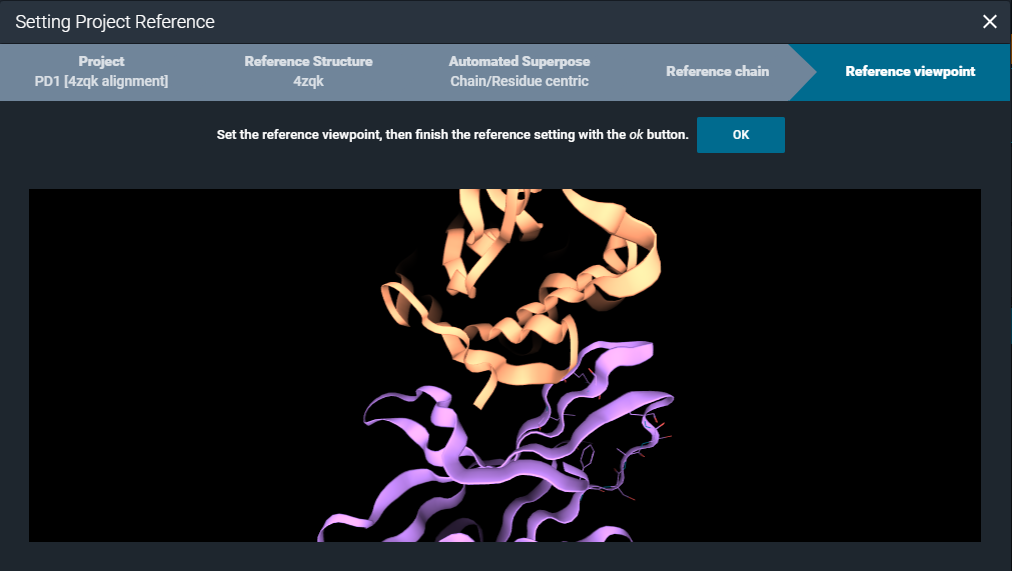
In this way, you have the collection of structures in the project (and any additional structures added to the project at a later time) pre-aligned to the reference protein chain or protein region of the project reference structure.