¶ Overview
The Pocket Similarity Search in 3decision enables you to search for similar binding sites across the entire 3decision knowledge base, including proprietary and public structures as well as AlphaFold models. This feature helps you discover structurally similar local environments in proteins, providing insights for multiple use cases:
- Discover new chemical matter by finding ligands that bind to similar pockets
- Identify potential off-targets for compound design and safety monitoring
- Find similar protein conformations across different structures
- Compare subpockets, not just entire binding sites—a unique capability that goes beyond traditional pocket comparison tools
The updated Pocket Similarity Search (3decision v2.3) features real-time results, improved accuracy, custom structure upload, and advanced filtering options to give you precise control over your searches.
This section of the User Guide describes the revamped and enhanced Pocket Similarity Search algorithm used in 3decision since version 2.3.
For previous 3decision versions, refer to this section of the documentation, that describes the previous Pocket Search algorithm.
Contents
- Accessing Pocket Similarity Search
- Search Options
- Working with Results
- Downloading Results
- Troubleshooting & FAQ
- How It Works (Background)
¶ Accessing Pocket Similarity Search
You can launch a Pocket Similarity Search in two ways:
- From a structure that is already registered in the 3decision database (from the Workspace)
 |
 |
|---|
- From a local structure file that you upload
 |
 |
|---|
¶ Method 1: From a registered structure
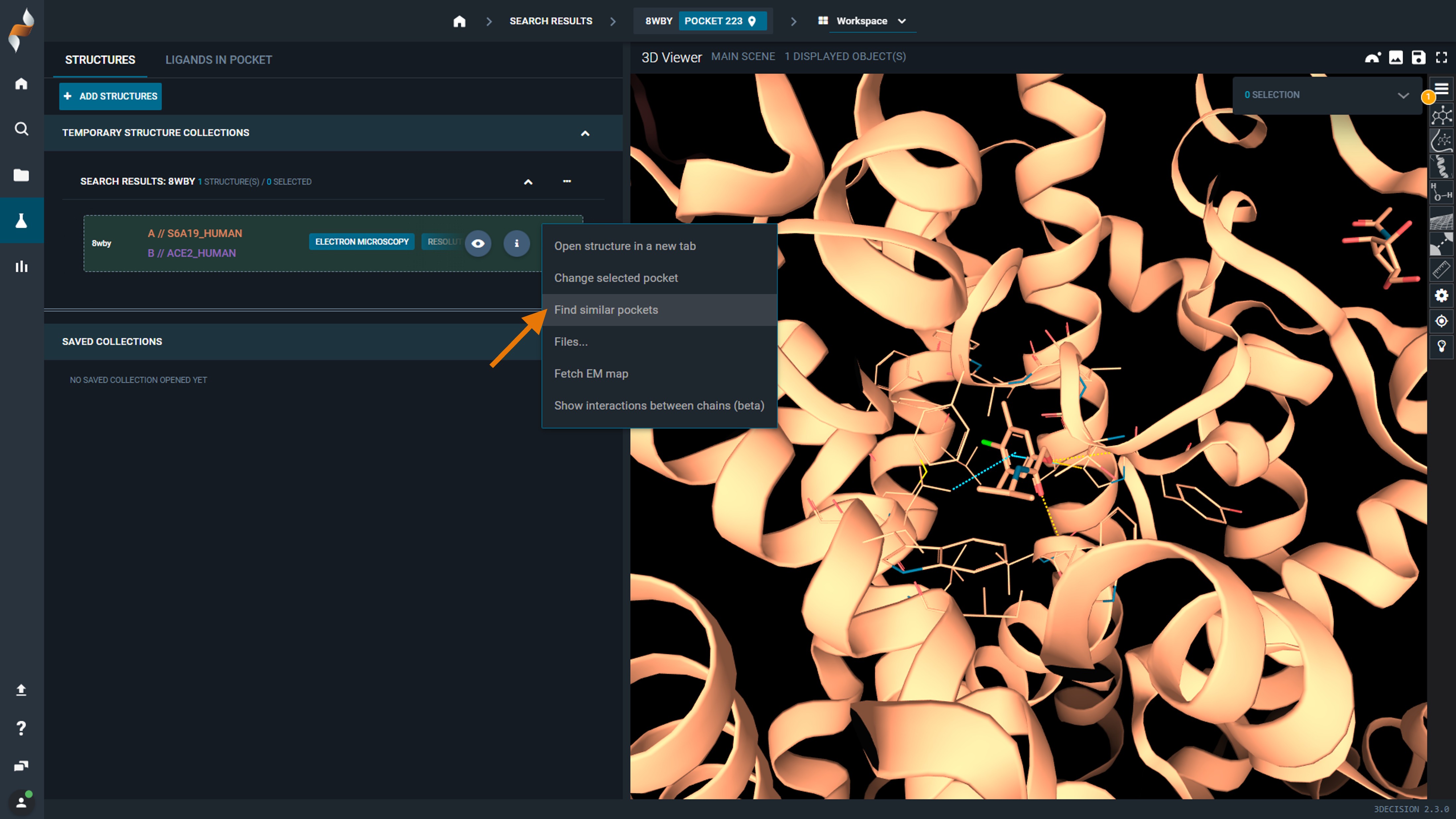
- Open your structure in the workspace
- Click the three-dot button (Structure Actions) on the structure line
 |
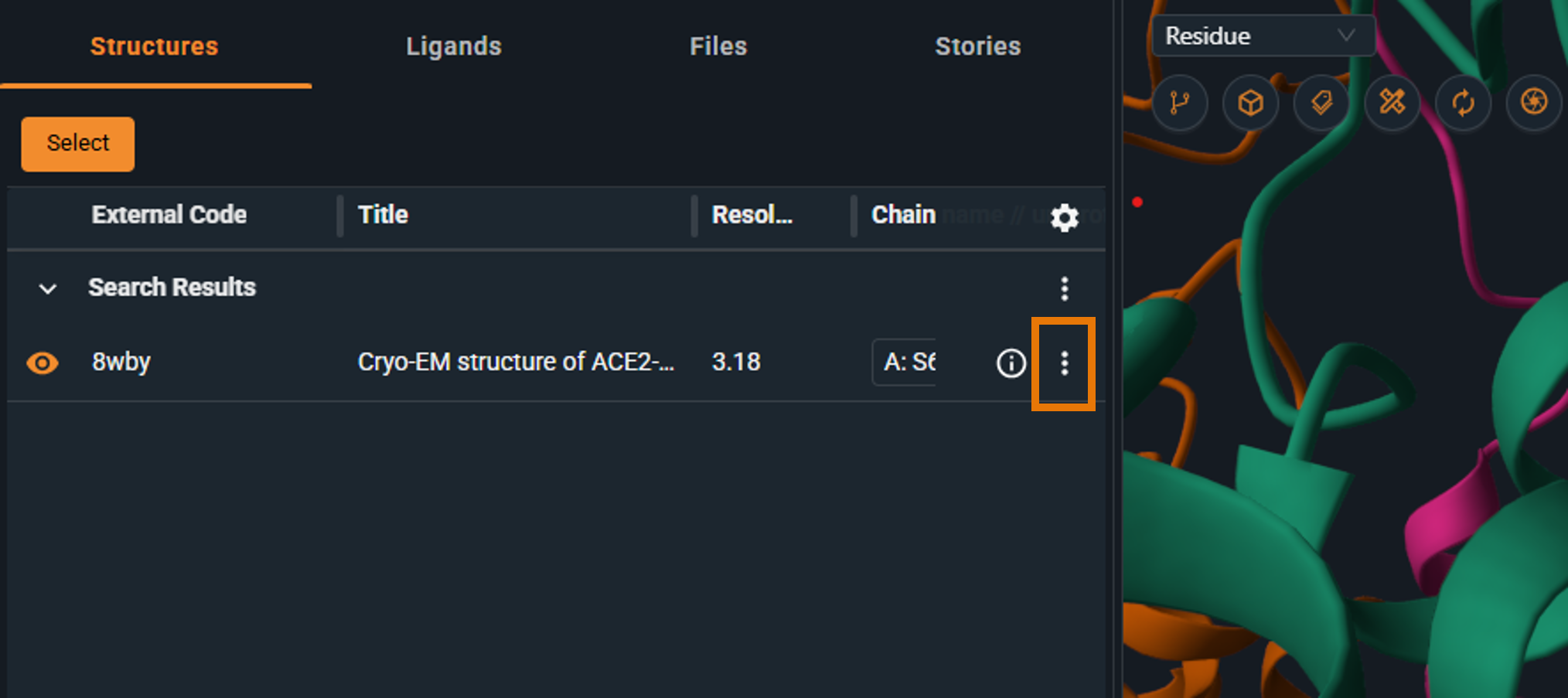 |
|---|
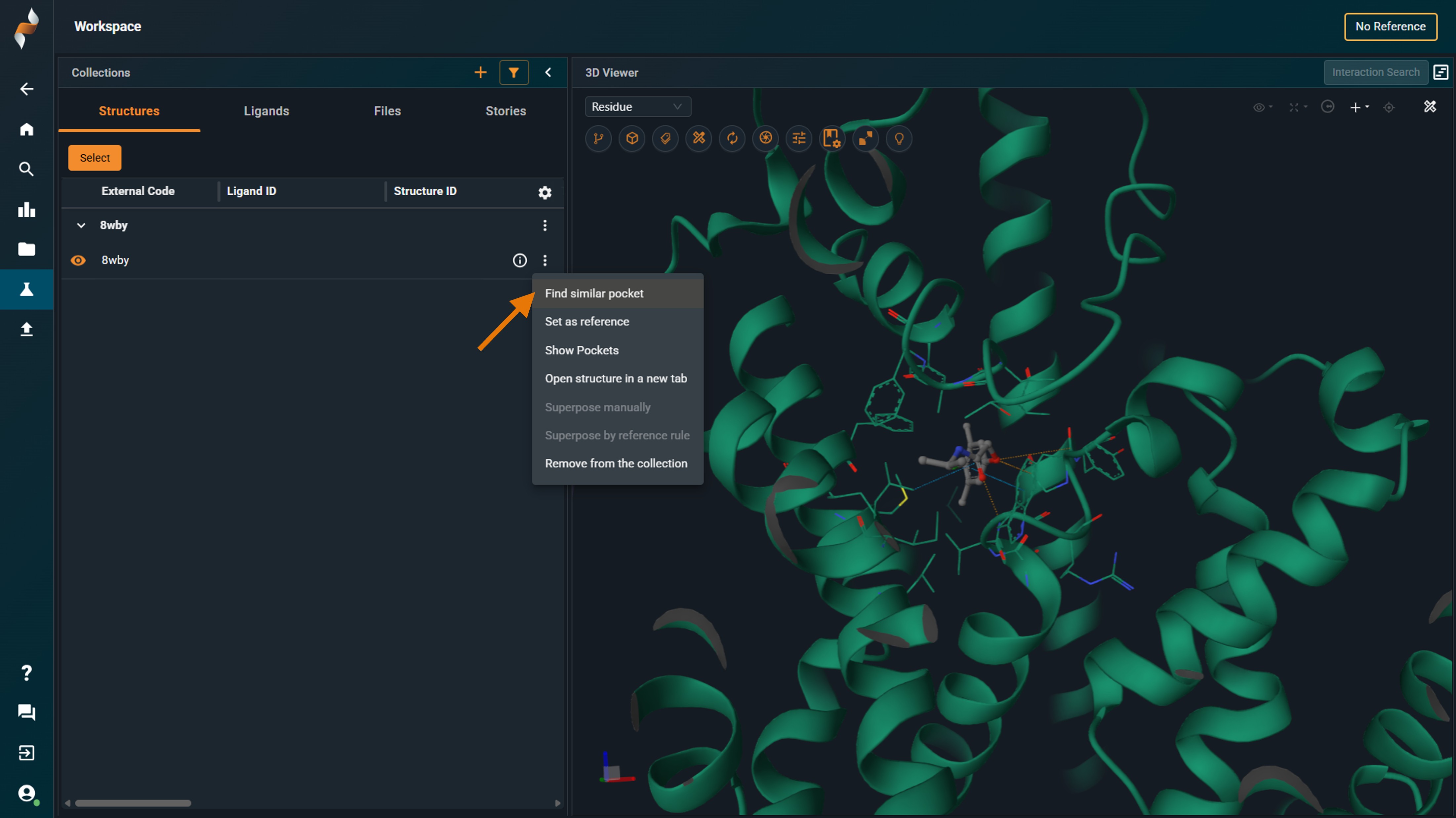
- Select "Find similar pockets"
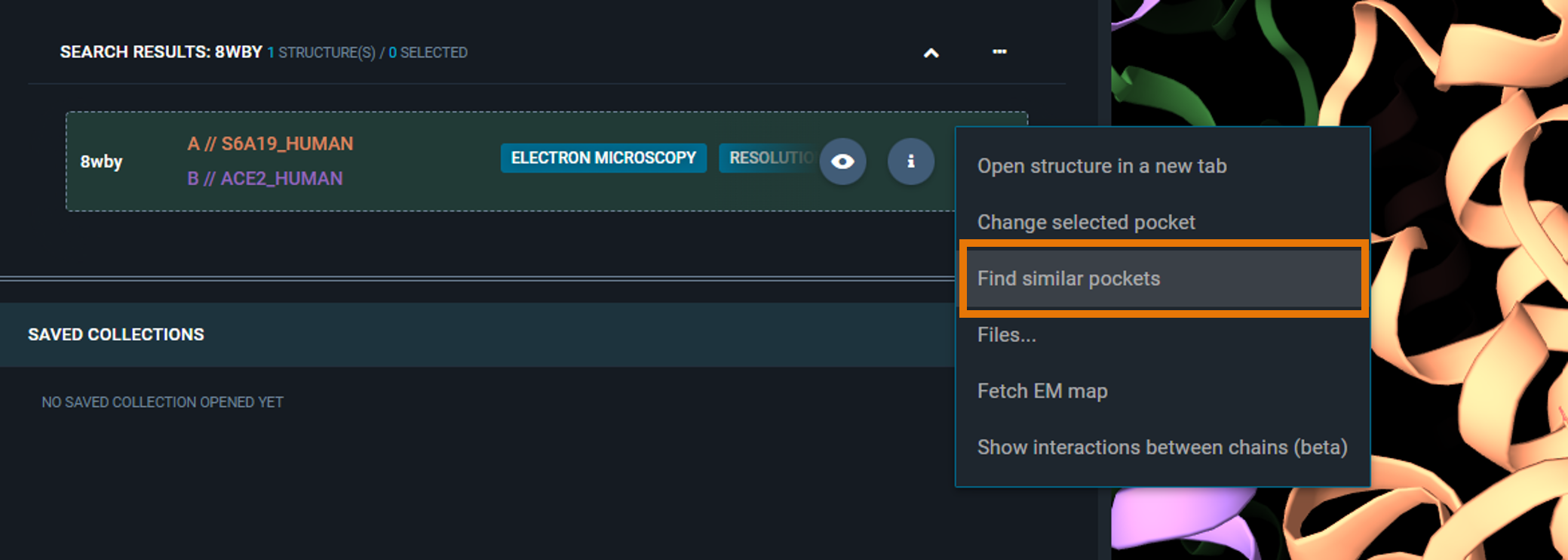 |
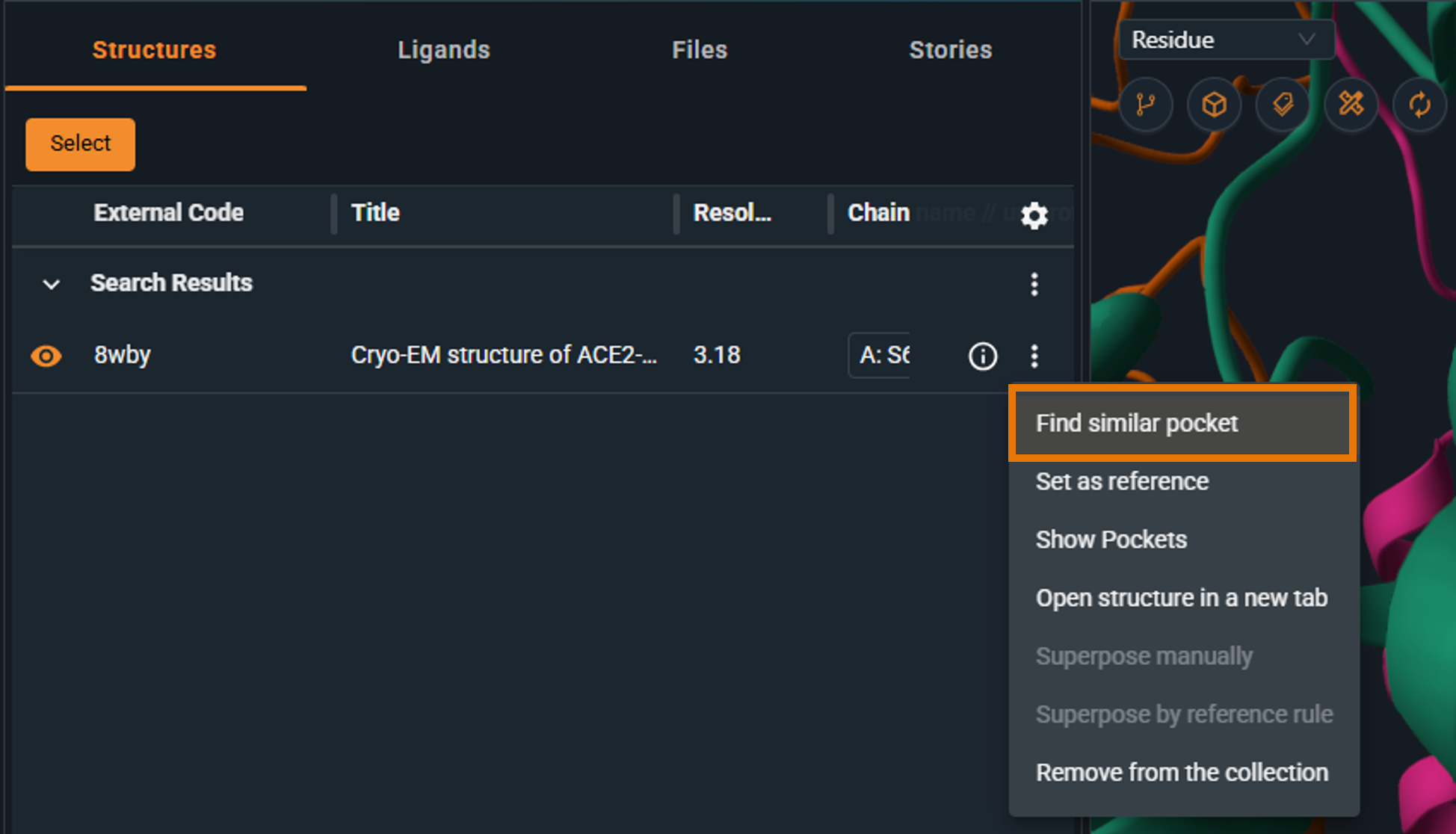 |
|---|
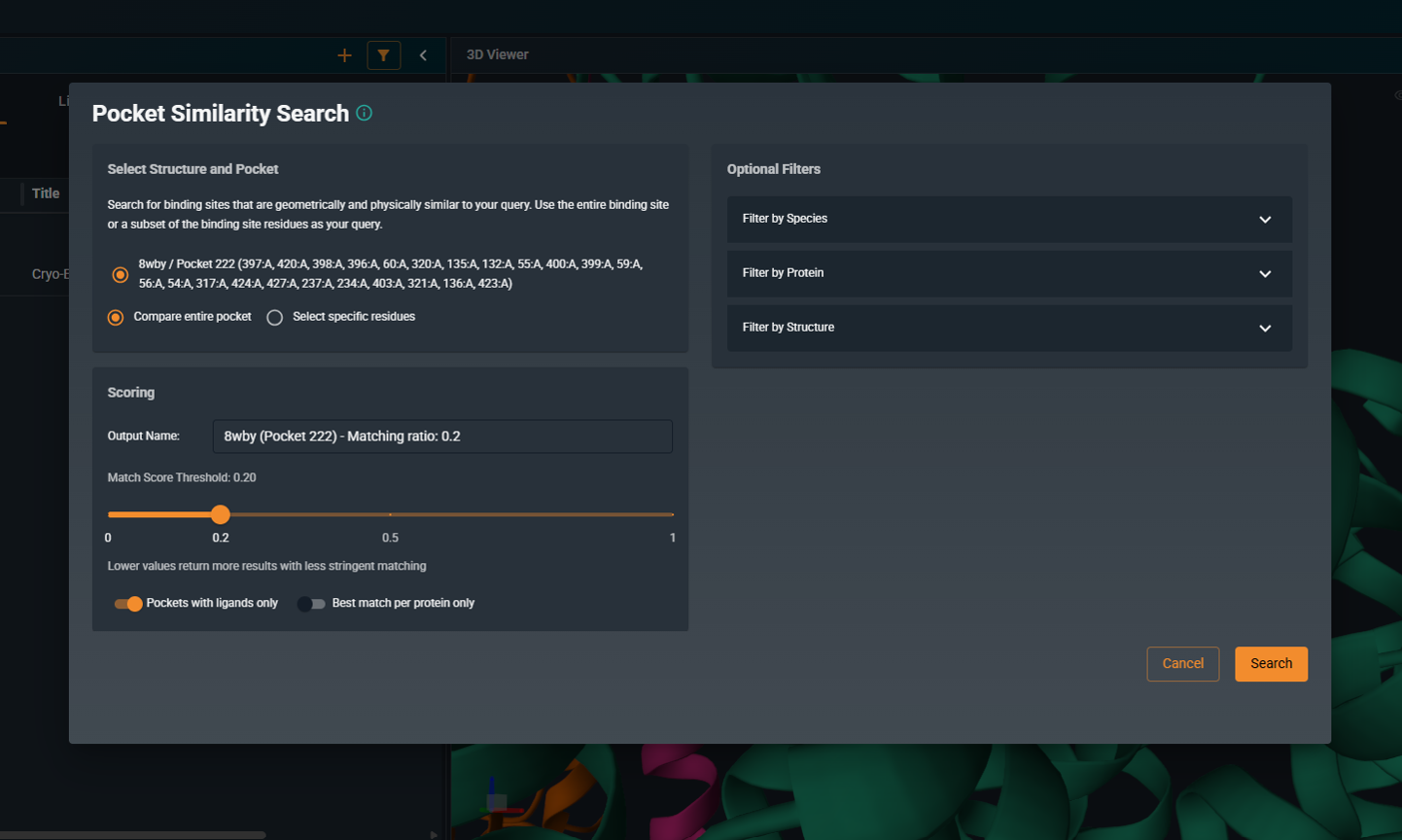
- Configure search parameters and filters as needed, then click SEARCH
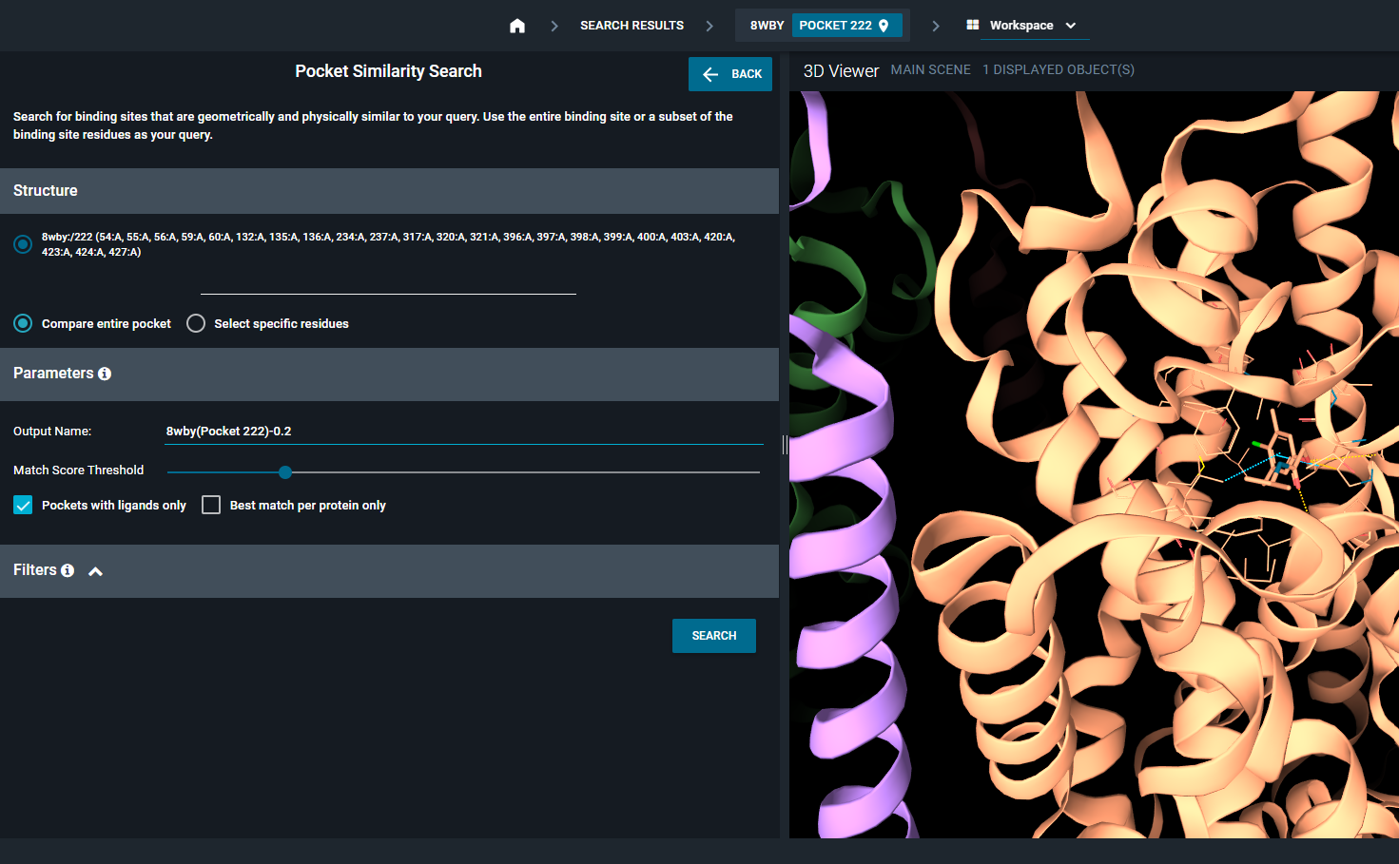 |
 |
|---|
A dedicated tutorial will be published shortly.
¶ Method 2: From a local file
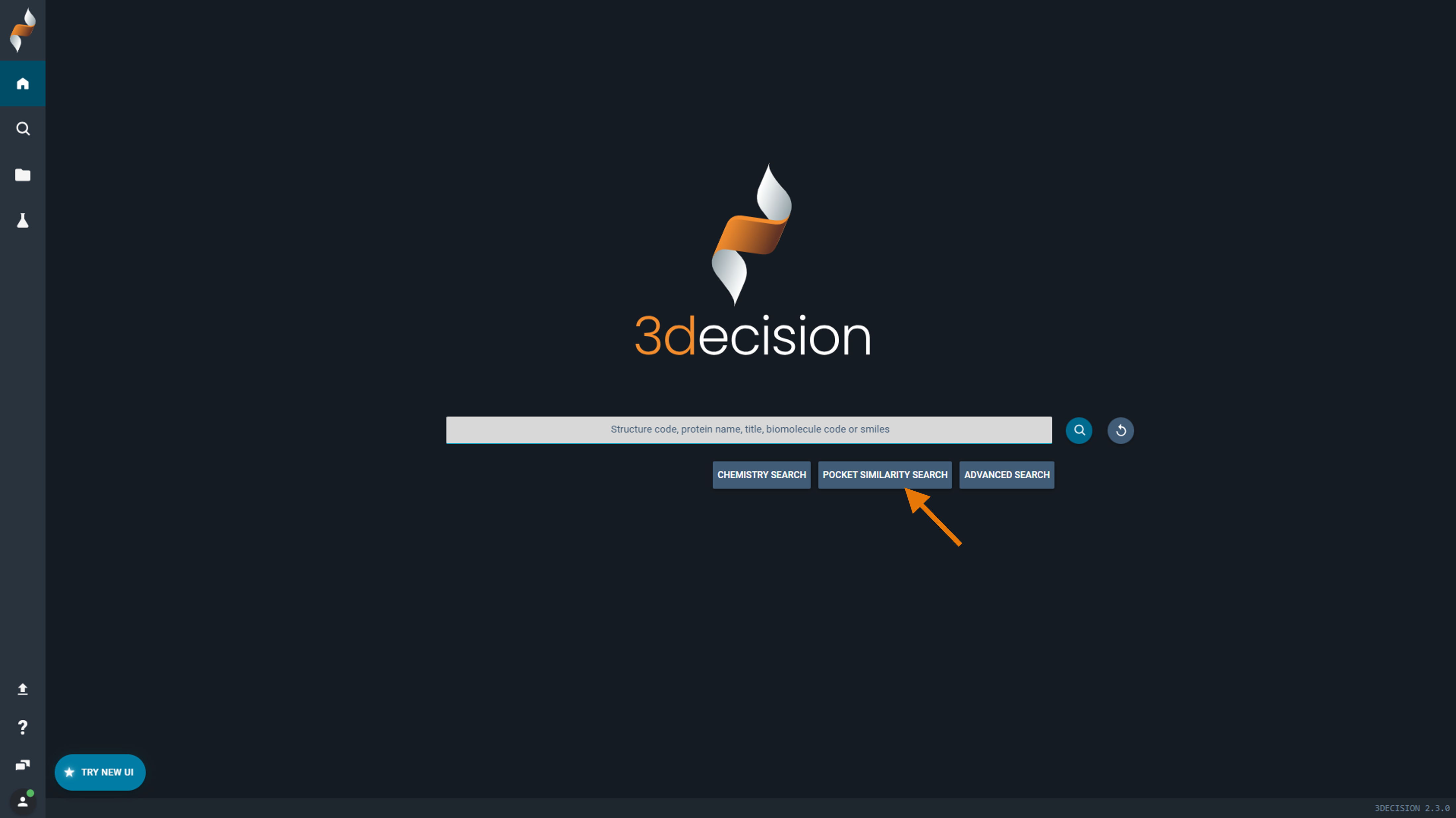
This option is available on the 3decision home page, directly under the search bar.
- On the home page, click Pocket Similarity Search (from file)
 |
 |
|---|
- Upload your structure file
 |
 |
|---|
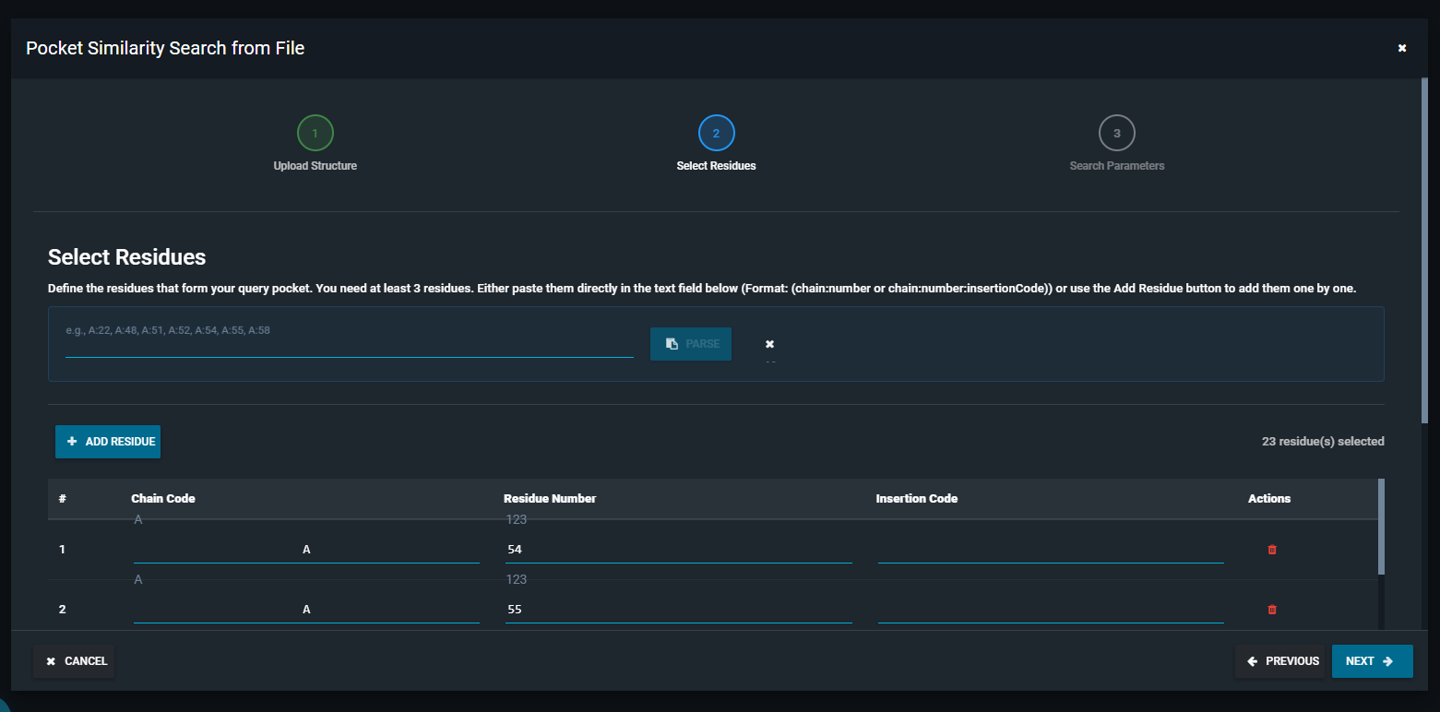
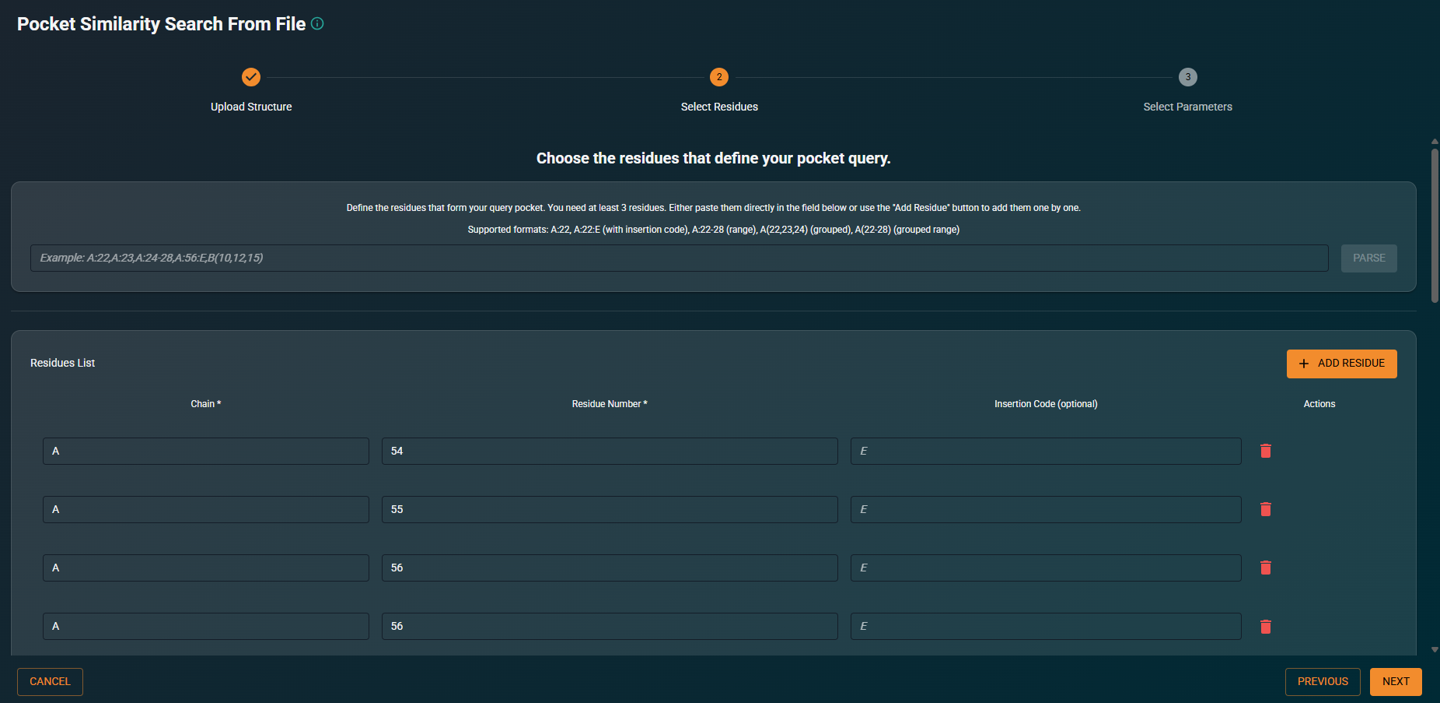
- Enter the list of residue numbers that define your query pocket
 |
 |
|---|
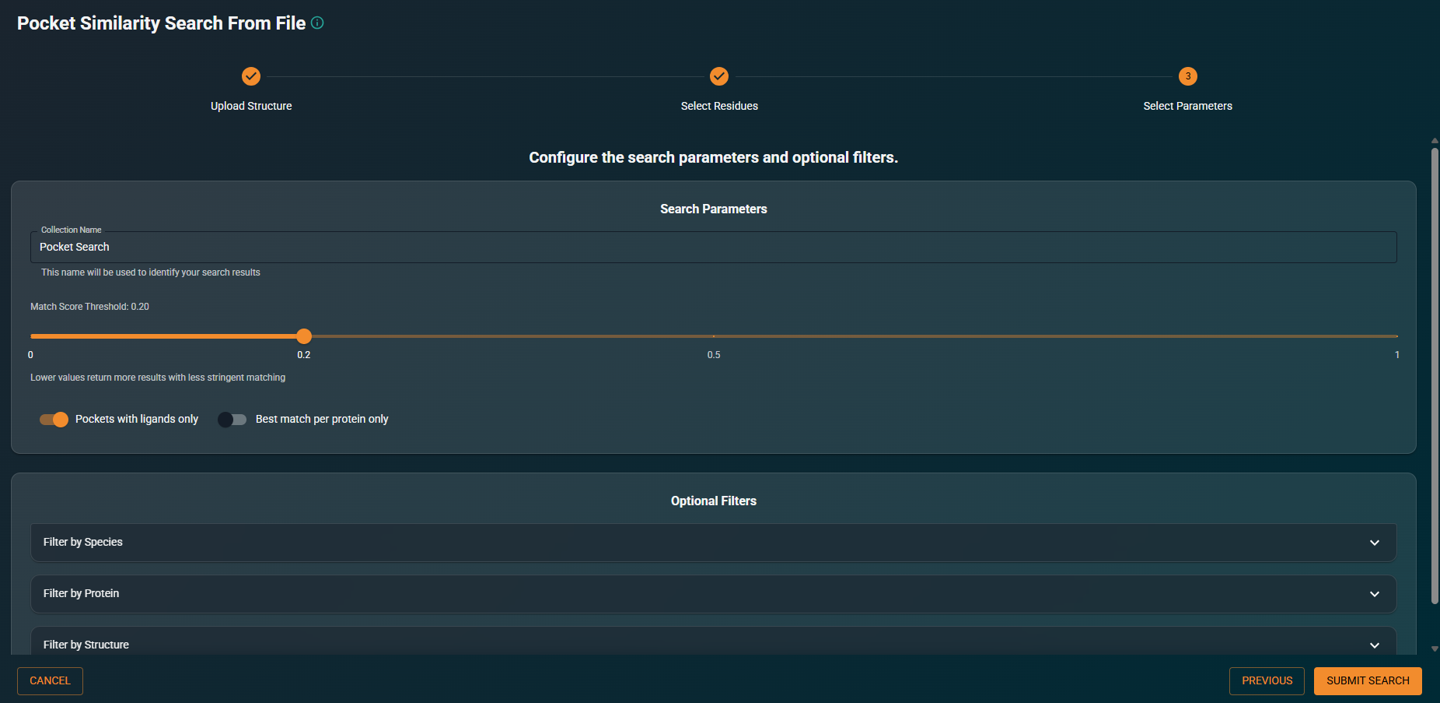
- Configure search parameters and filters as needed, then click SEARCH
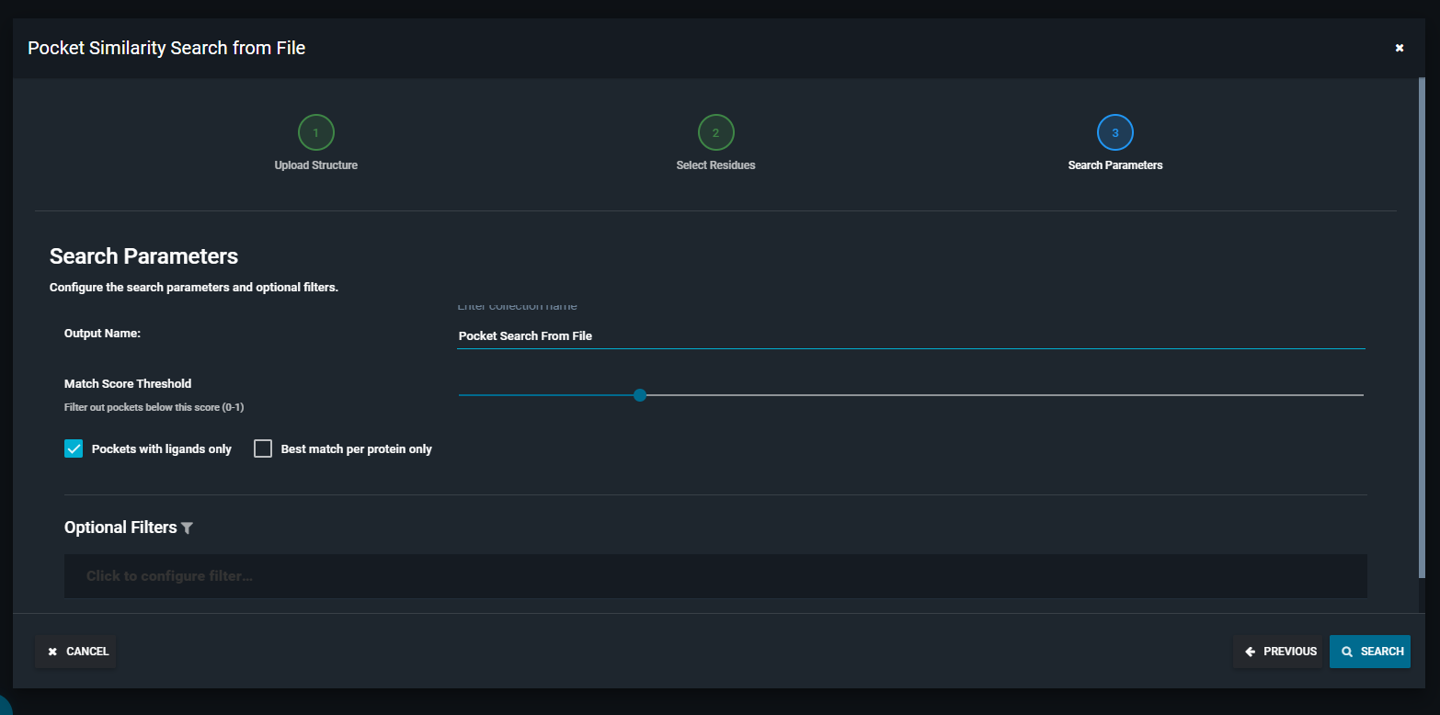 |
 |
|---|
A dedicated tutorial will be published shortly.
¶ Search Options
¶ Pocket definition
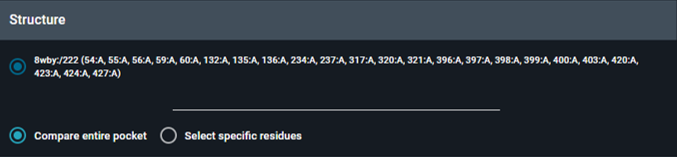
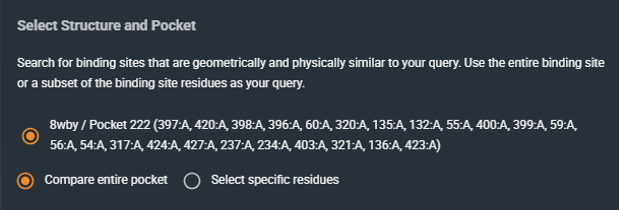
Choose how the query pocket is defined:
-
Compare entire pocket: Uses all residues lining the selected pocket (full pocket search).
-
Select specific residues: Uses only the residues you select (subpocket search). This is useful when you want to focus on a specific region or interaction motif.
-
Default: Compare entire pocket (full pocket)
 |
 |
|---|
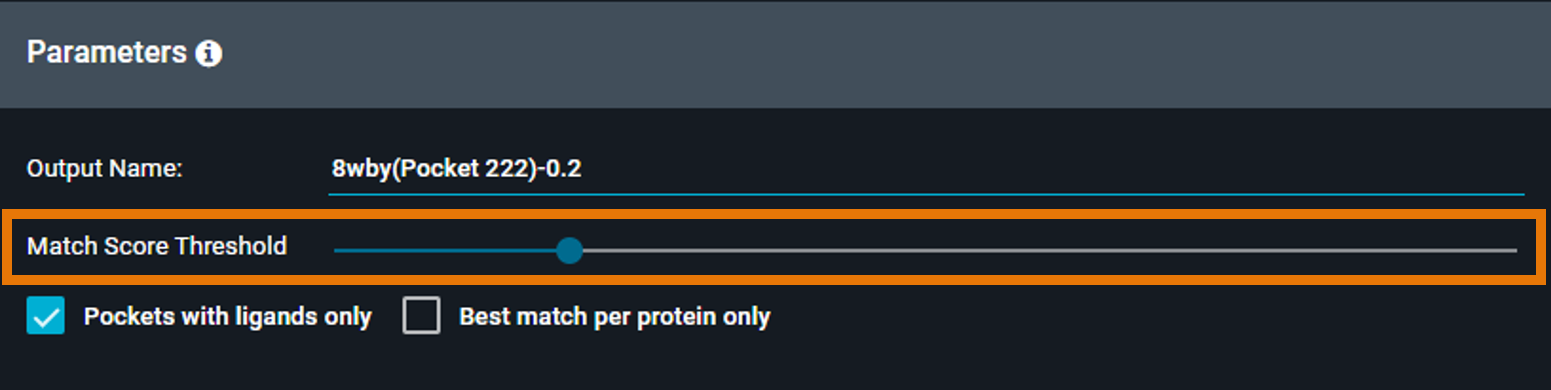
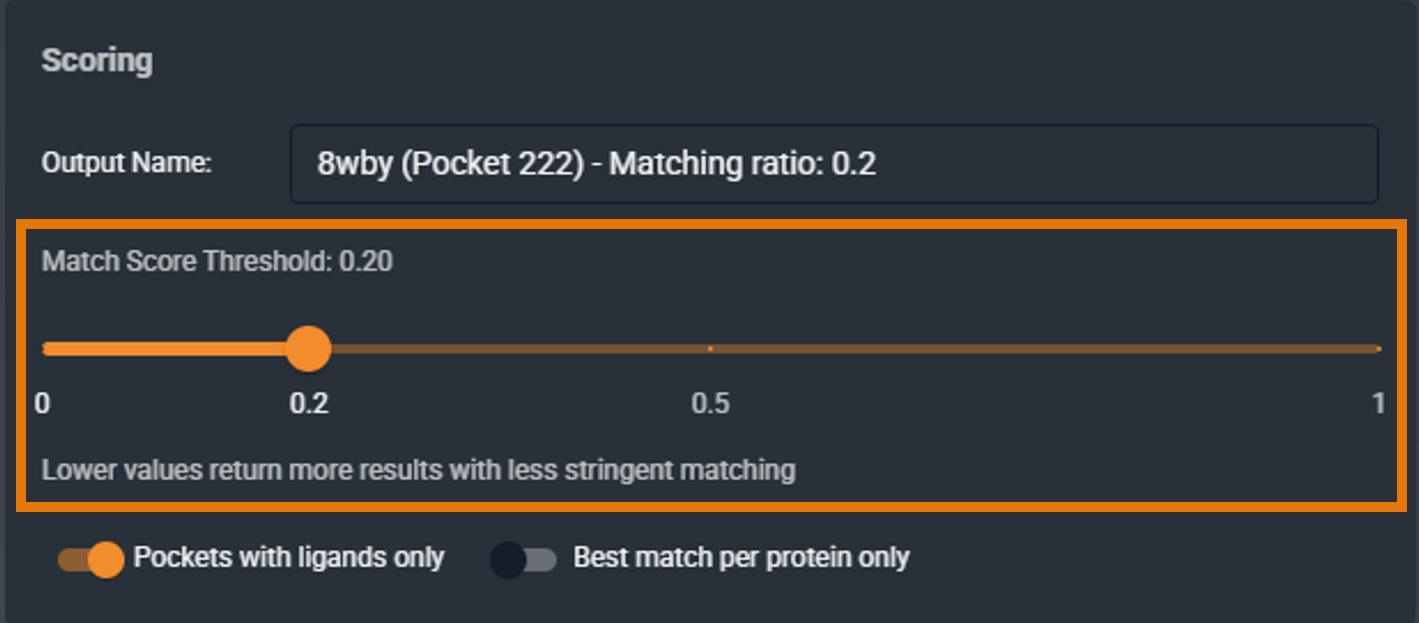
¶ Score Threshold
The score threshold determines which matches are returned. Only pockets with a similarity score at or above this threshold will appear in your results.
- Default: 0.2
- Range: 0.0 to 1.0
 |
 |
|---|
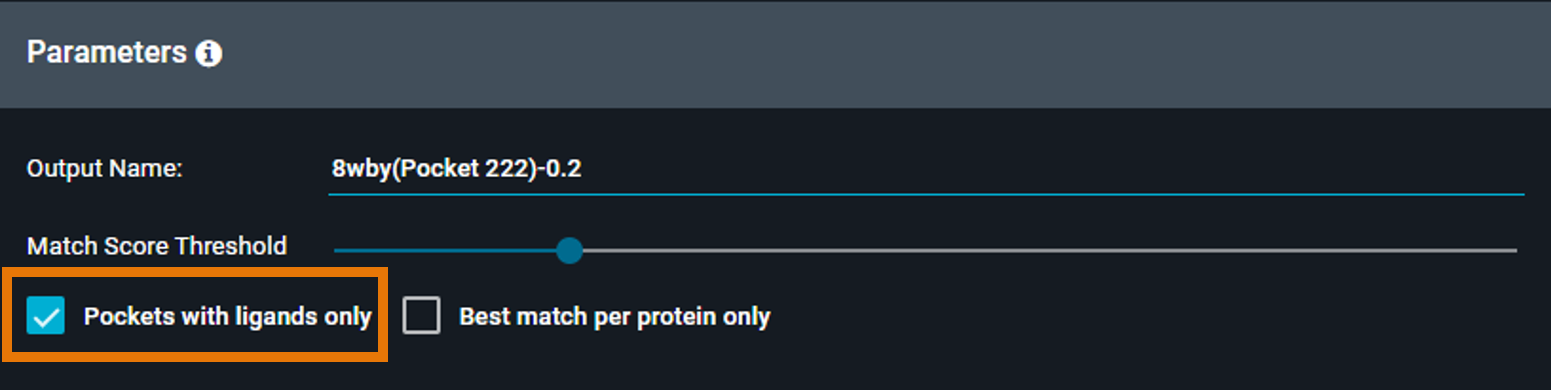
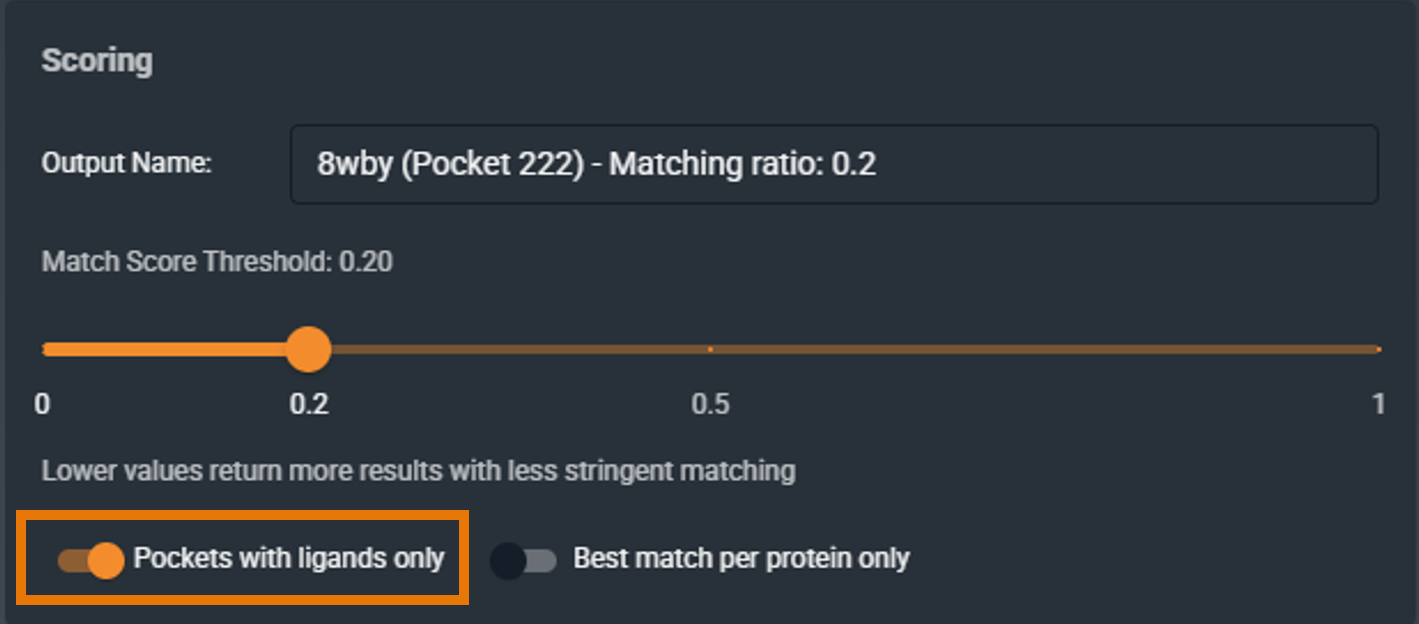
¶ Pockets with Ligands Only
When enabled, the search only includes binding sites that contain a ligand (holo structures).
-
Checked: Find chemical matter that binds to similar pockets
-
Unchecked: Search the full database including apo structures, useful for off-target prediction
-
Default: Checked
 |
 |
|---|
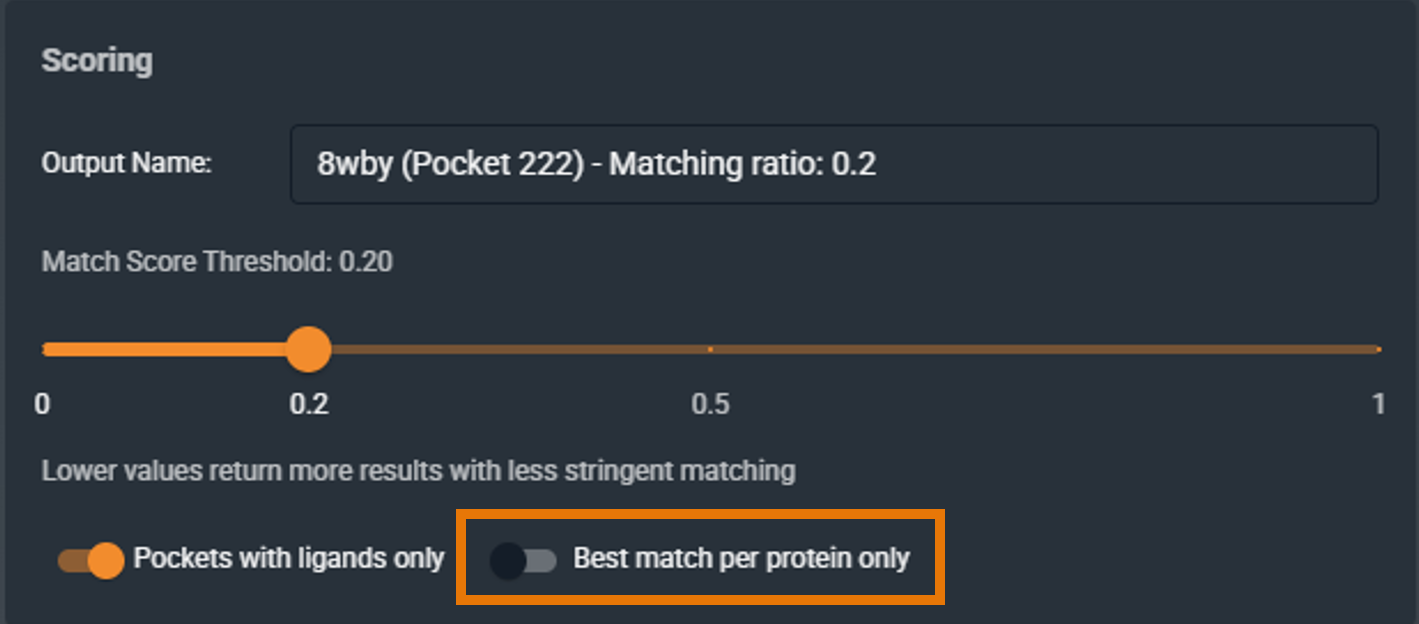
¶ Best Match per Protein Only
When enabled, returns only one representative structure per protein, reducing redundancy.
- Checked: Useful when you have many structures of the same target and want to focus on cross-target relationships
- Unchecked: See all conformational variants and individual structures
- Default: Unchecked
 |
 |
|---|
¶ Filtering Options
Use filters to narrow your search to specific subsets of the database.
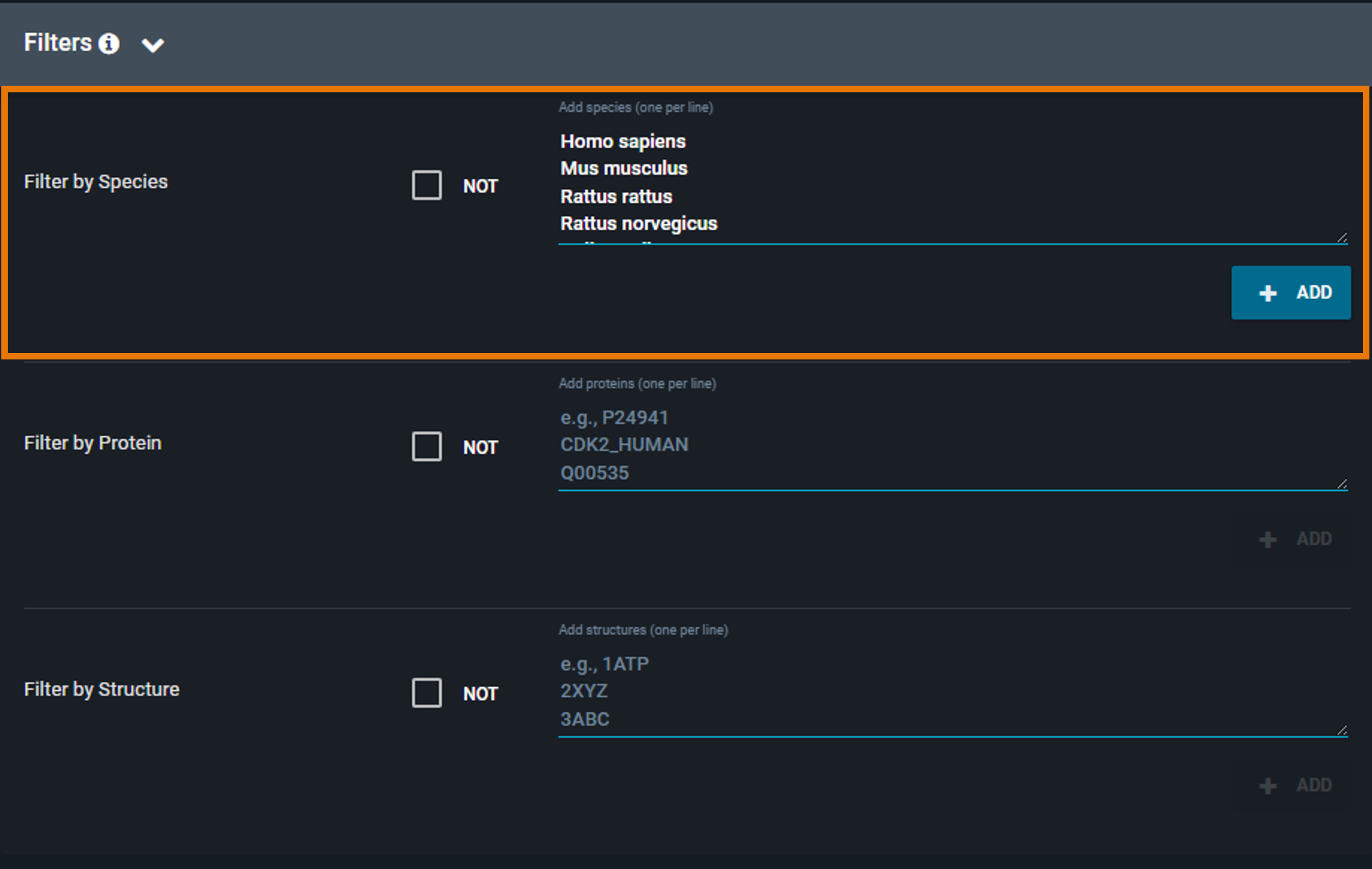
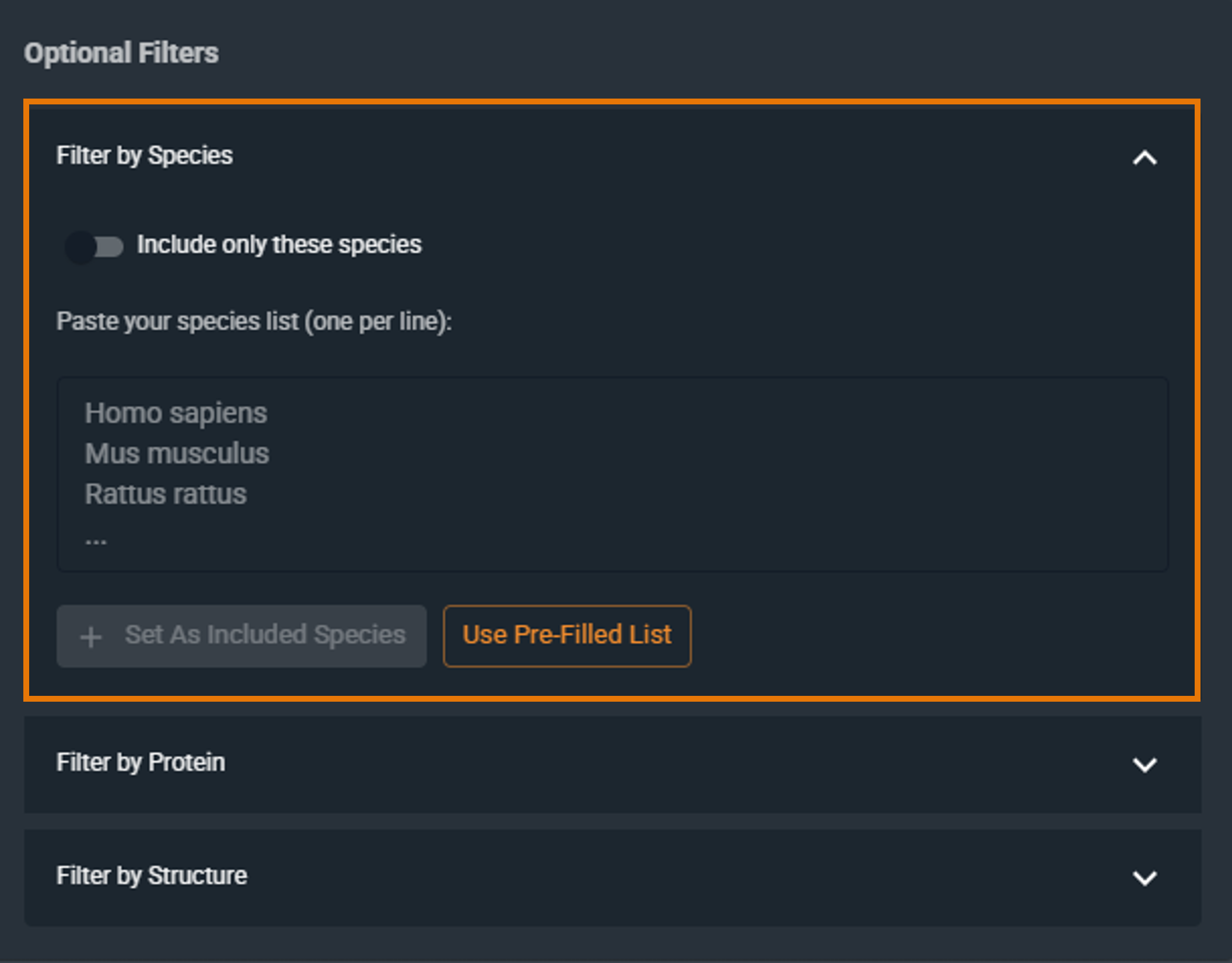
¶ Filter by Species
Restrict the search to specific organisms:
- Click Use Pre-Filled List and choose one or more species, OR
- Enter the Latin species name manually (one species per line) in the text box, then click Set As Included Species.
To remove a species, click the X next to it.
By default, the filter includes the selected species. To exclude them instead, click the Include only these species checkbox to switch to Exclude these species (the checkbox is located above the filter text box).
 |
 |
|---|
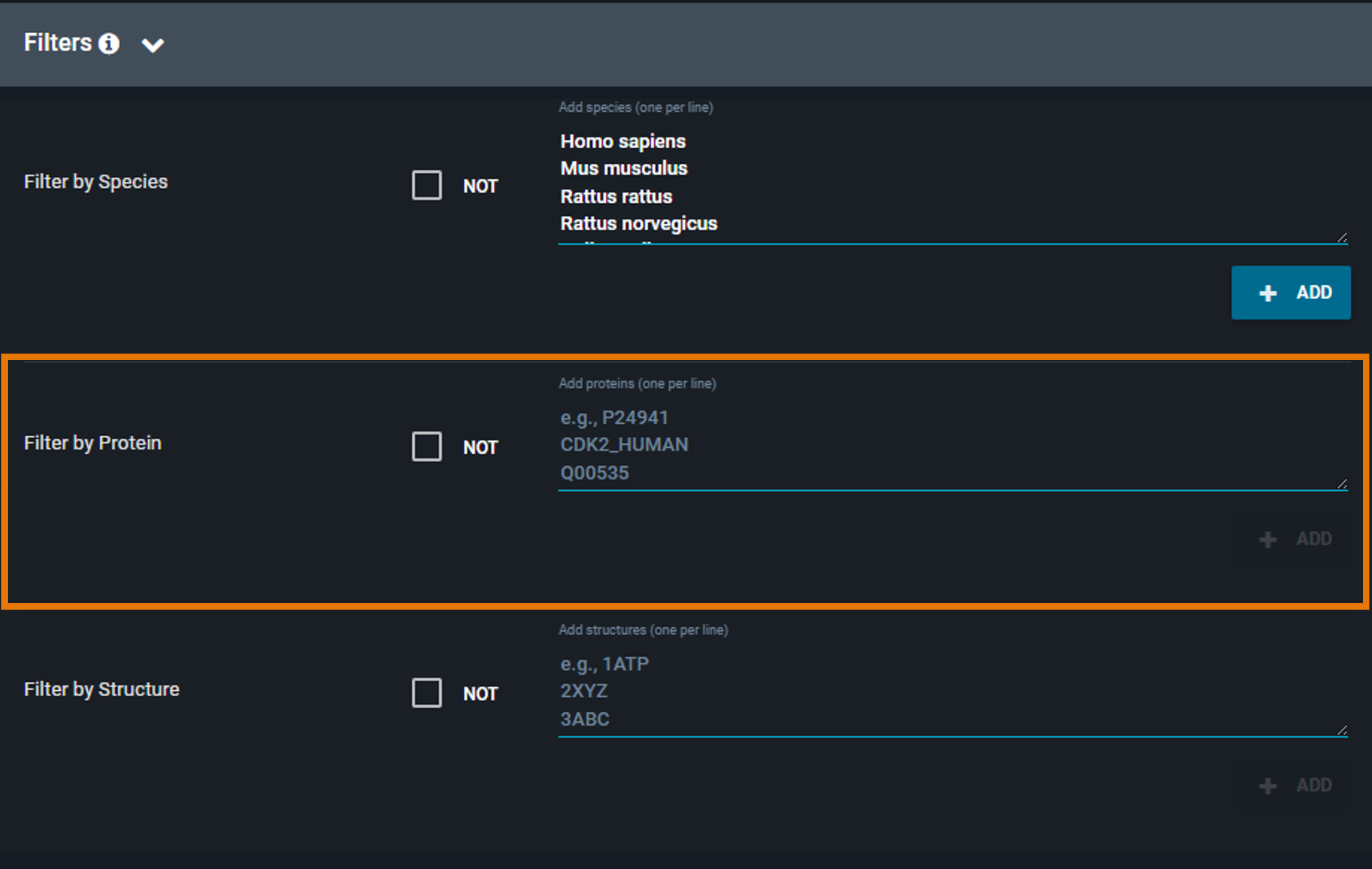
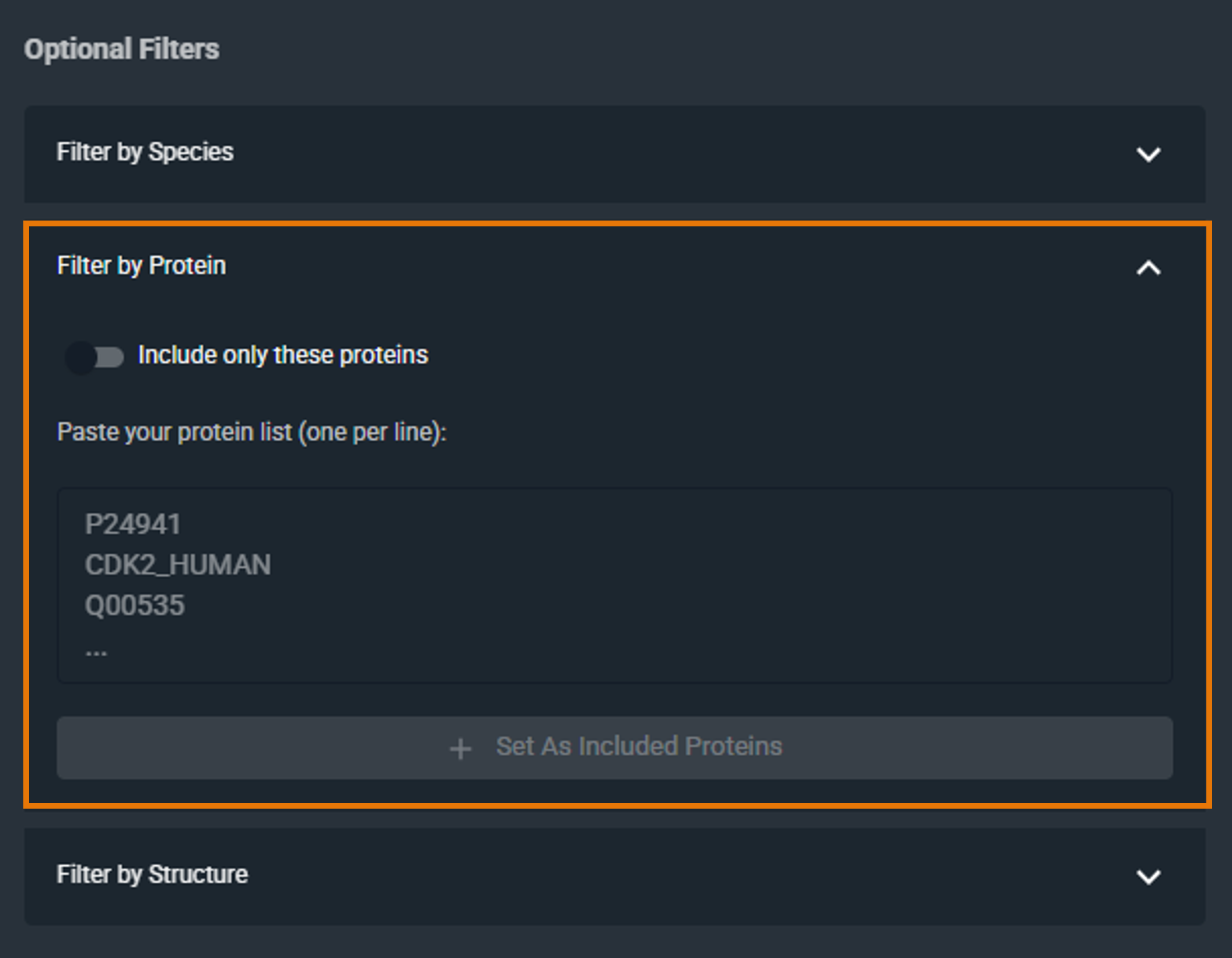
¶ Filter by Protein
Restrict the search to specific proteins by specifying a list of UniProt identifiers or accession codes:
- Enter one or more UniProt IDs or accession codes (one per line)
- Click Set As Included Proteins to include them in your filter
To remove a protein, click the X next to it.
By default, the filter includes the selected proteins. To exclude them instead, click the Include only these proteins checkbox to switch to Exclude these proteins (the checkbox is located above the filter text box).
 |
 |
|---|
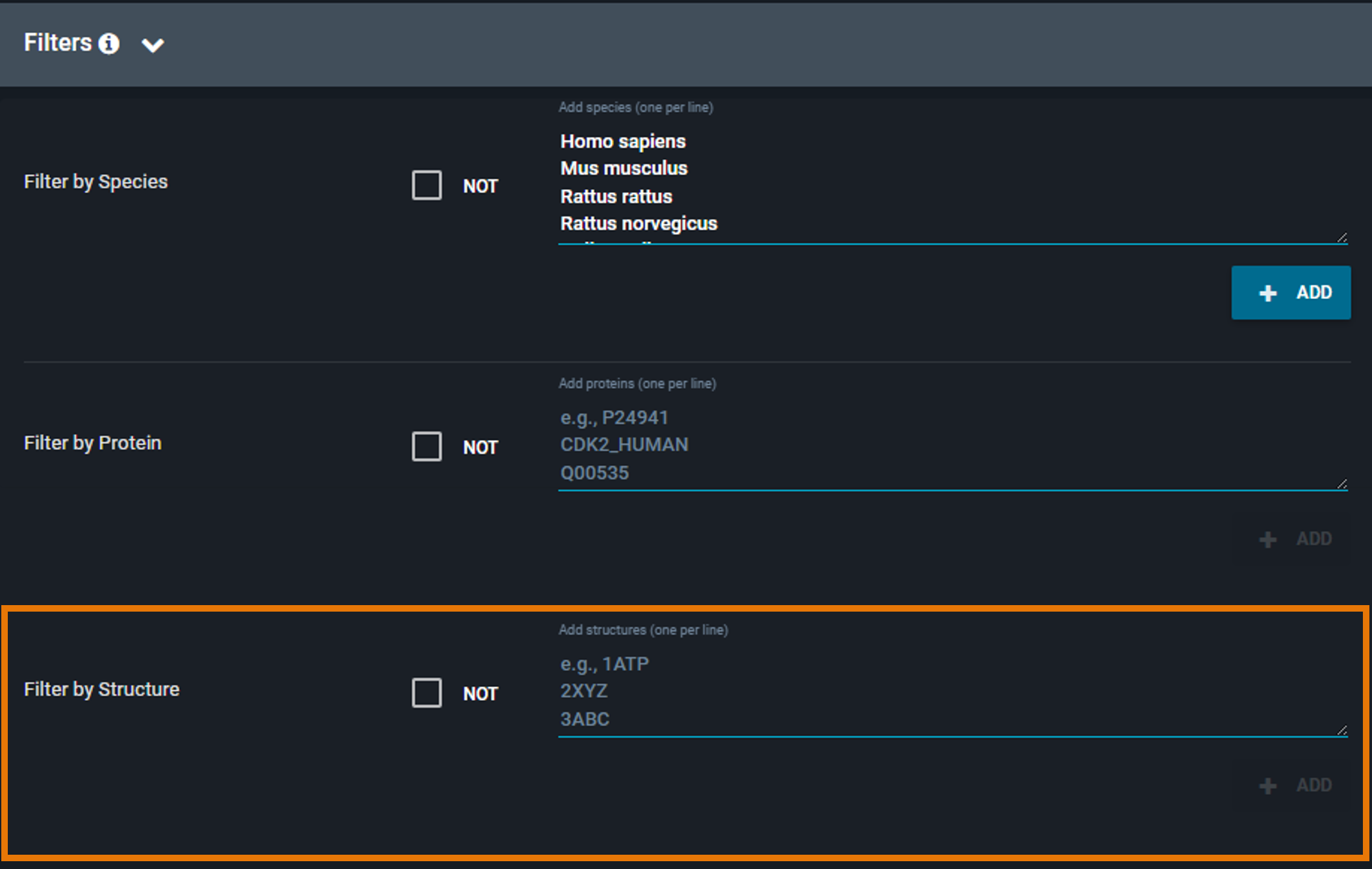
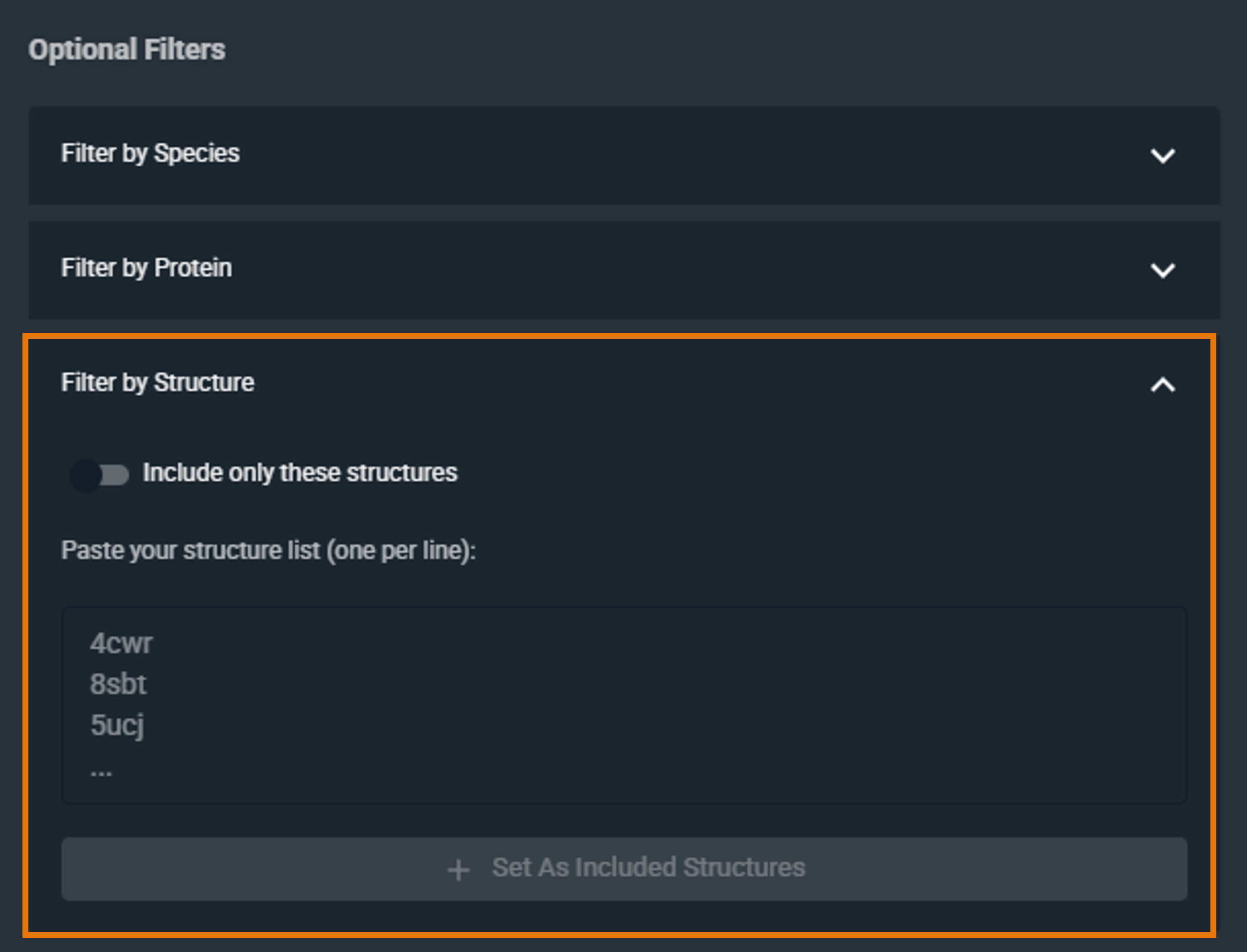
¶ Filter by PDB
Restrict the search to a specific set of structures by providing a list of PDB codes:
- Enter one or more PDB codes or 3decision structure codes (i.e. external code), one per line
- Click Set As Included Structures to include them in your filter
To remove a PDB code, remove it from the list of added codes.
By default, the filter includes the selected structures. To exclude them instead, click the Include only these structures checkbox to switch to Exclude these structures (the checkbox is located above the filter text box).
 |
 |
|---|
¶ Working with Results
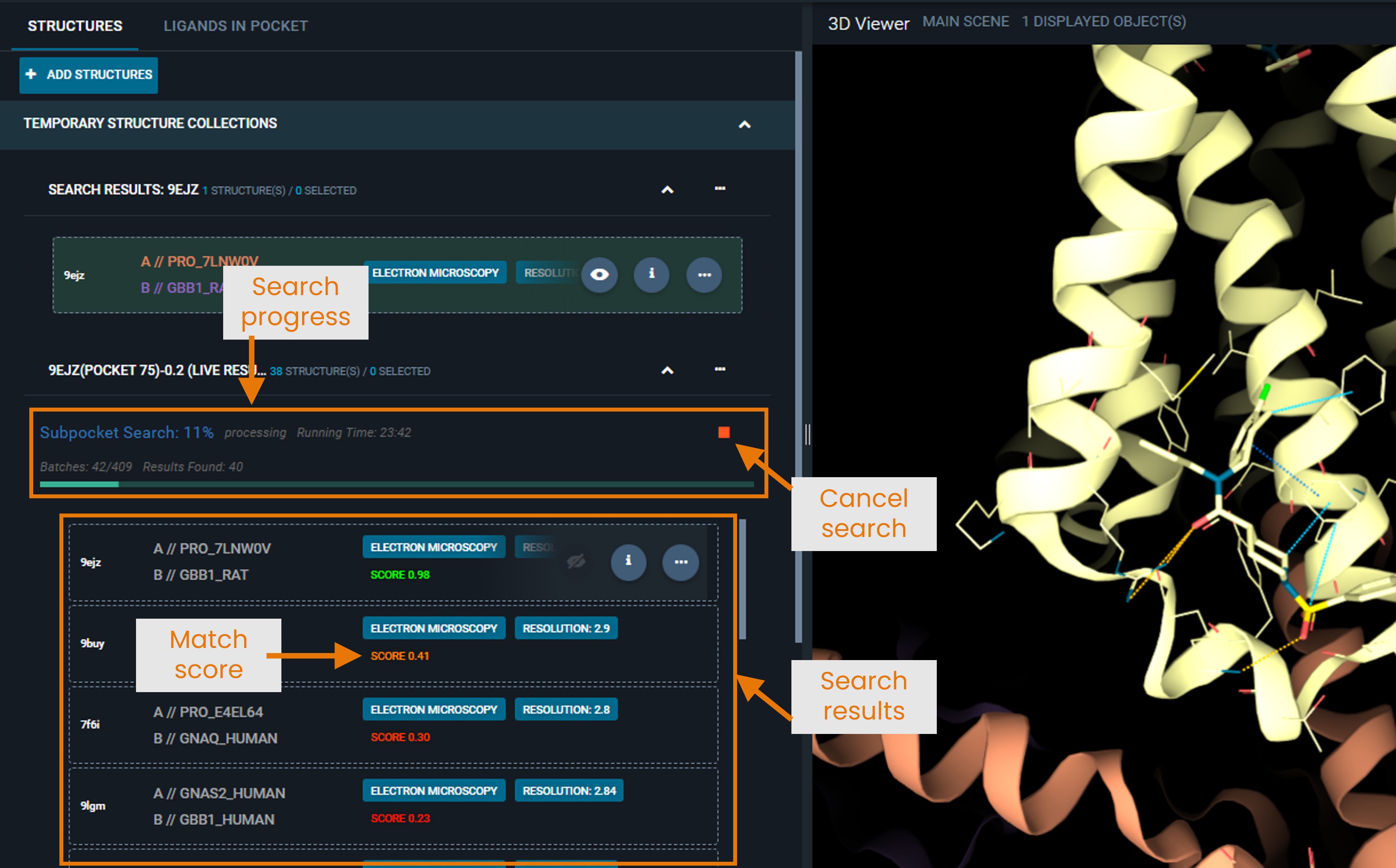
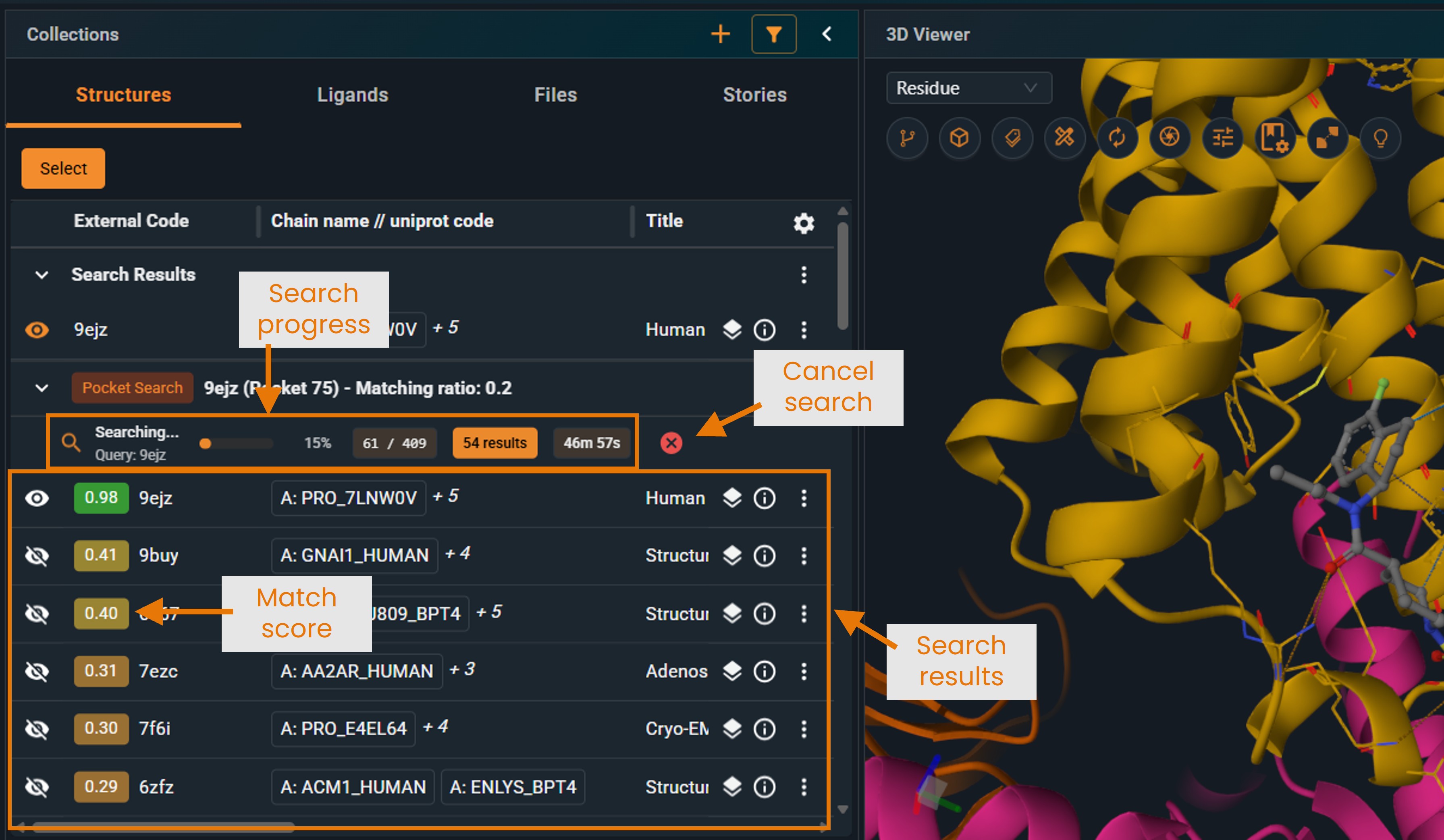
¶ Understanding the Results Panel
When you click SEARCH, the search begins running in the background:
- The system processes the search in batches (the feature-pair table is split into multiple batches)
- Progress bar shows how many batches have been processed
- Results appear in real-time as each batch completes
- You can stop the search at any time using the Cancel button
- Results are interactive even while the search continues
 |
 |
|---|
For large searches against the full production database, the search may take several tens of minutes. However, hits are retrieved and displayed in real time, so you can start analyzing results immediately without waiting for the search to complete. Note that the first hits displayed are not necessarily the highest-scoring ones; rankings may change as additional batches finish.
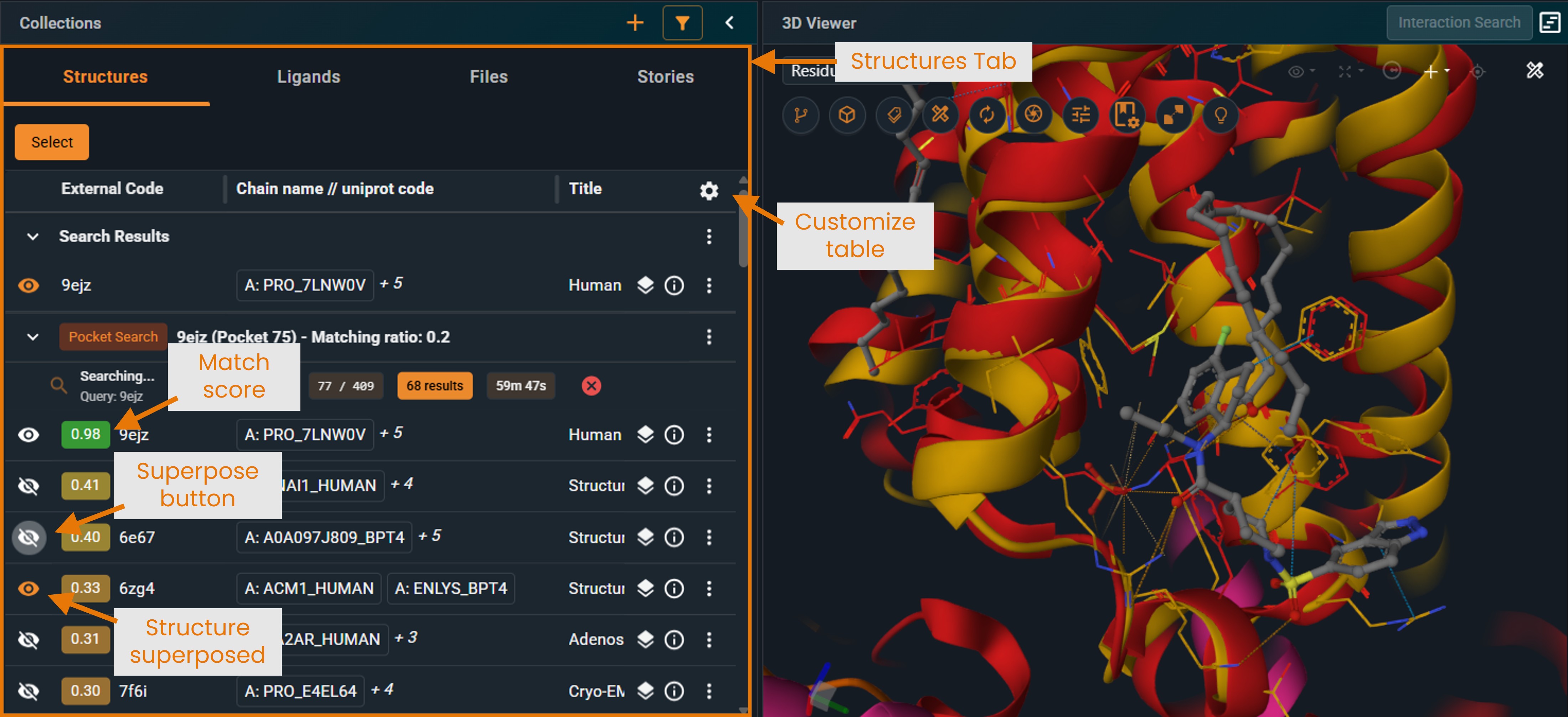
¶ Structure View Mode
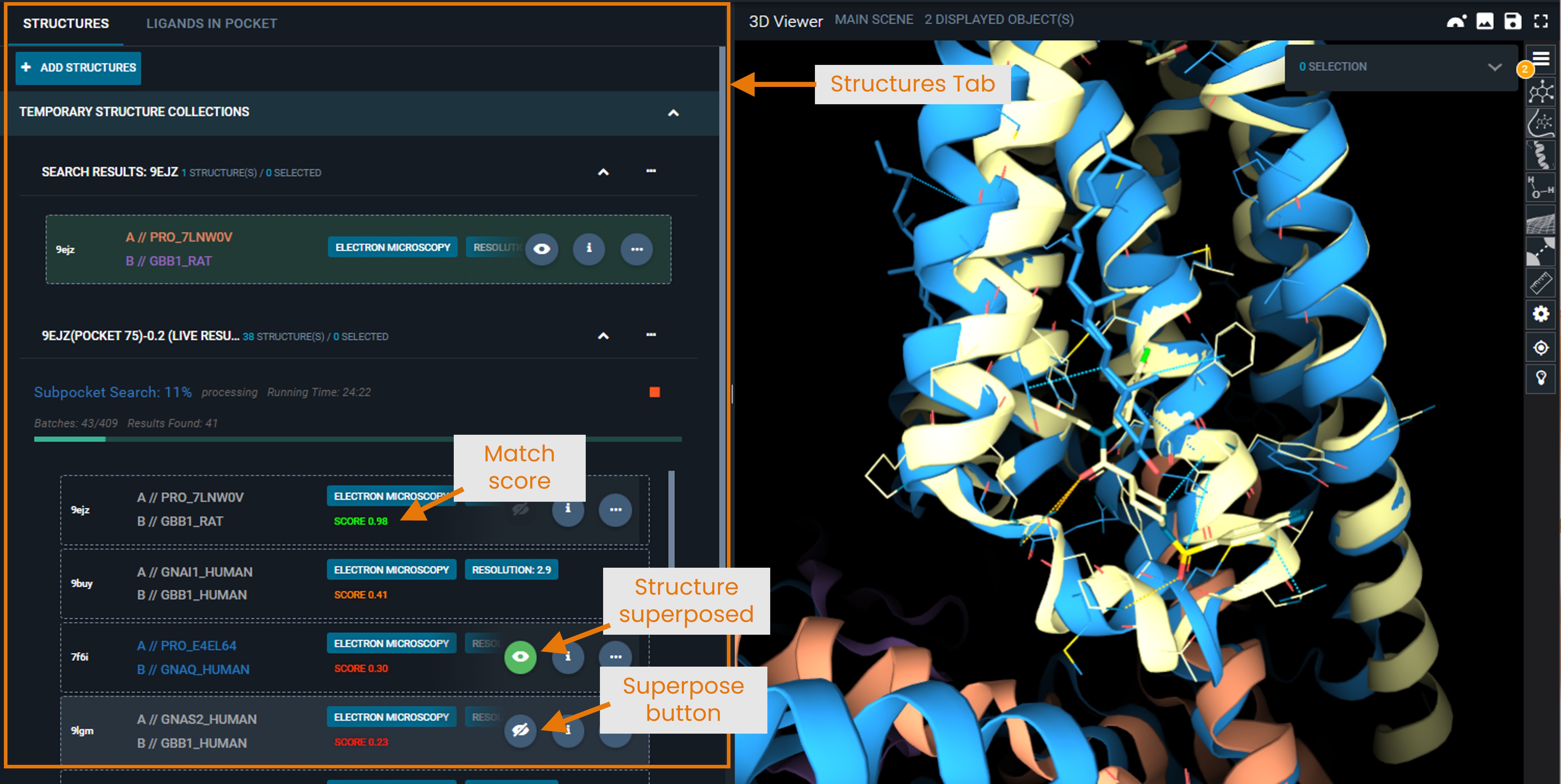
Results appear in a new collection in your Workspace under the Structure tab:
- Structures are sorted by match score (highest first)
- Each entry shows the structure name and similarity score.
- [New UI only] You can customize the table view to display other properties (e.g., UniProt name) by clicking the cogwheel at the top right of the panel.
- Click the eye icon to superpose structures for visual comparison. Superposition is computed as a distance-weighted fit on the Cα positions of the matched pocket residues (best cluster). To understand what a cluster is, see “Pocket Feature Clustering” in the “How Matching Works” section.
 |
 |
|---|
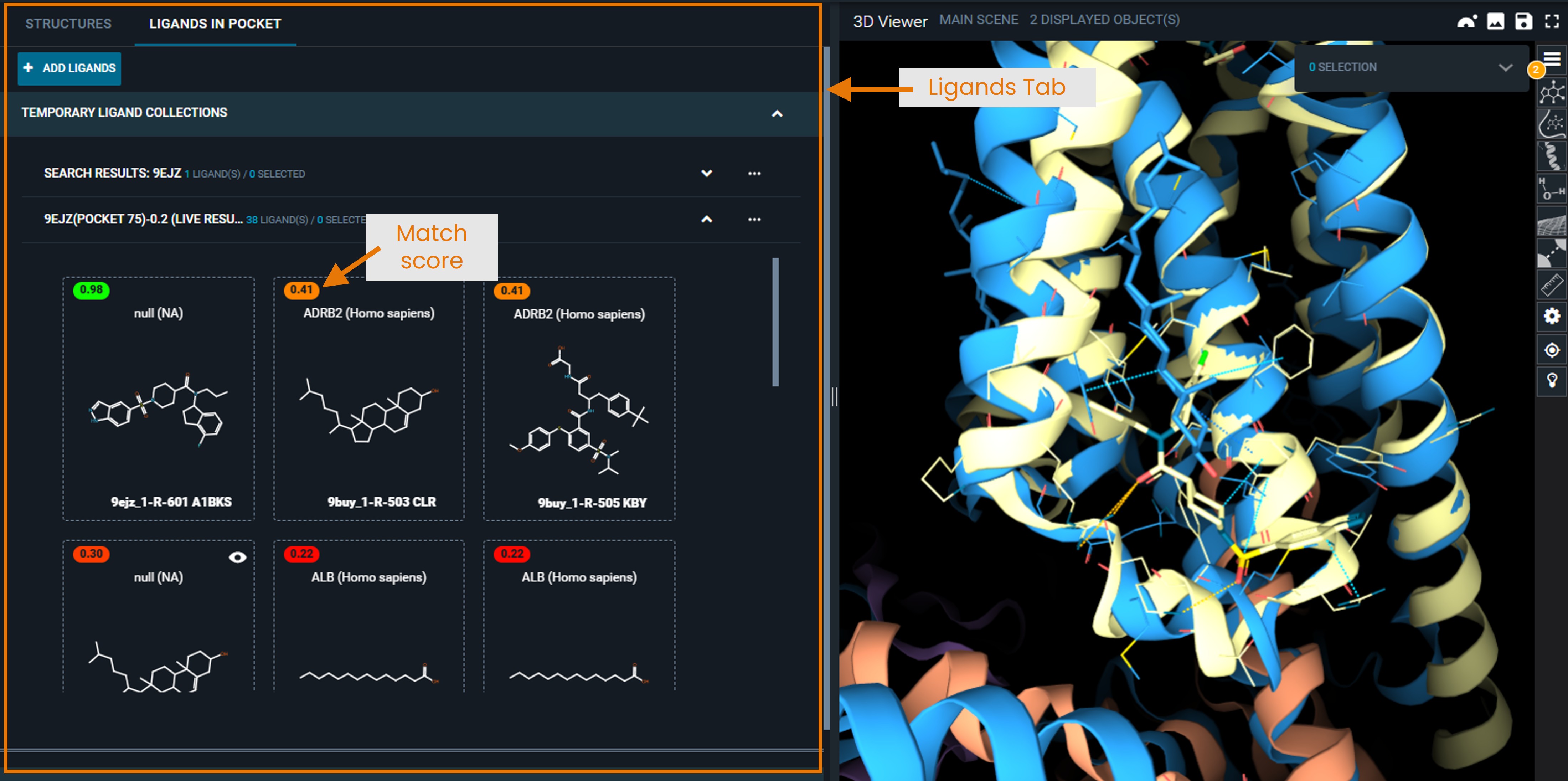
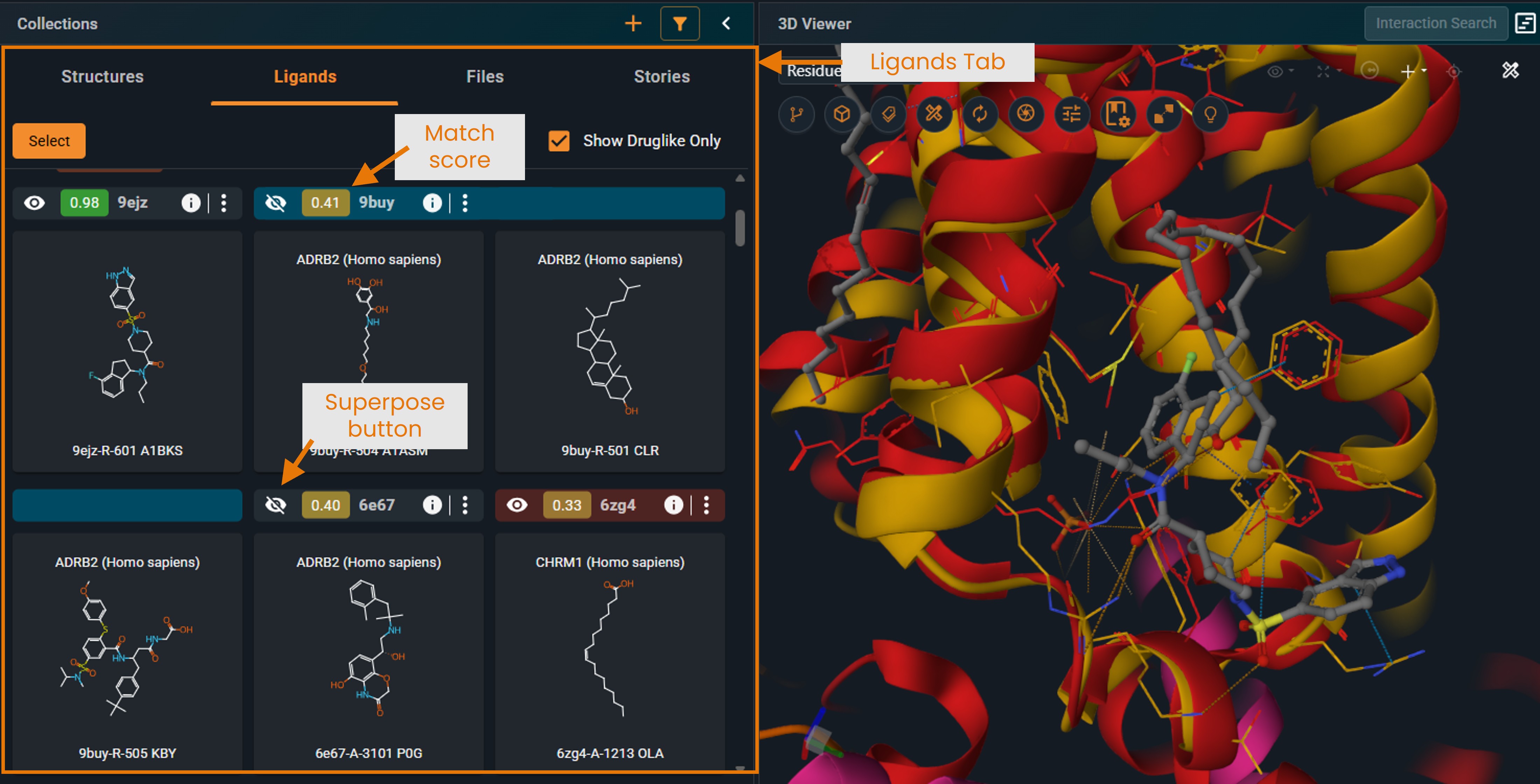
¶ Ligand View Mode
Switch to LIGANDS tab to display the ligands found in each structure:
- In the current version, all ligands found in the hit structure are displayed, not only the ligand located in the matching pocket. This will be improved in an upcoming version.
- Click the eye icon to superpose structures in the same way as in the Structure tab.
 |
 |
|---|
¶ Downloading Results
You can download either the full result collection or a selection of hits.
The steps for exporting are slightly different between the two versions of the User Interface, so are described separately below.
¶ New Beta UI
Download all results
- Open the action menu of the Pocket Similarity Search result collection.
- Select Download collection.
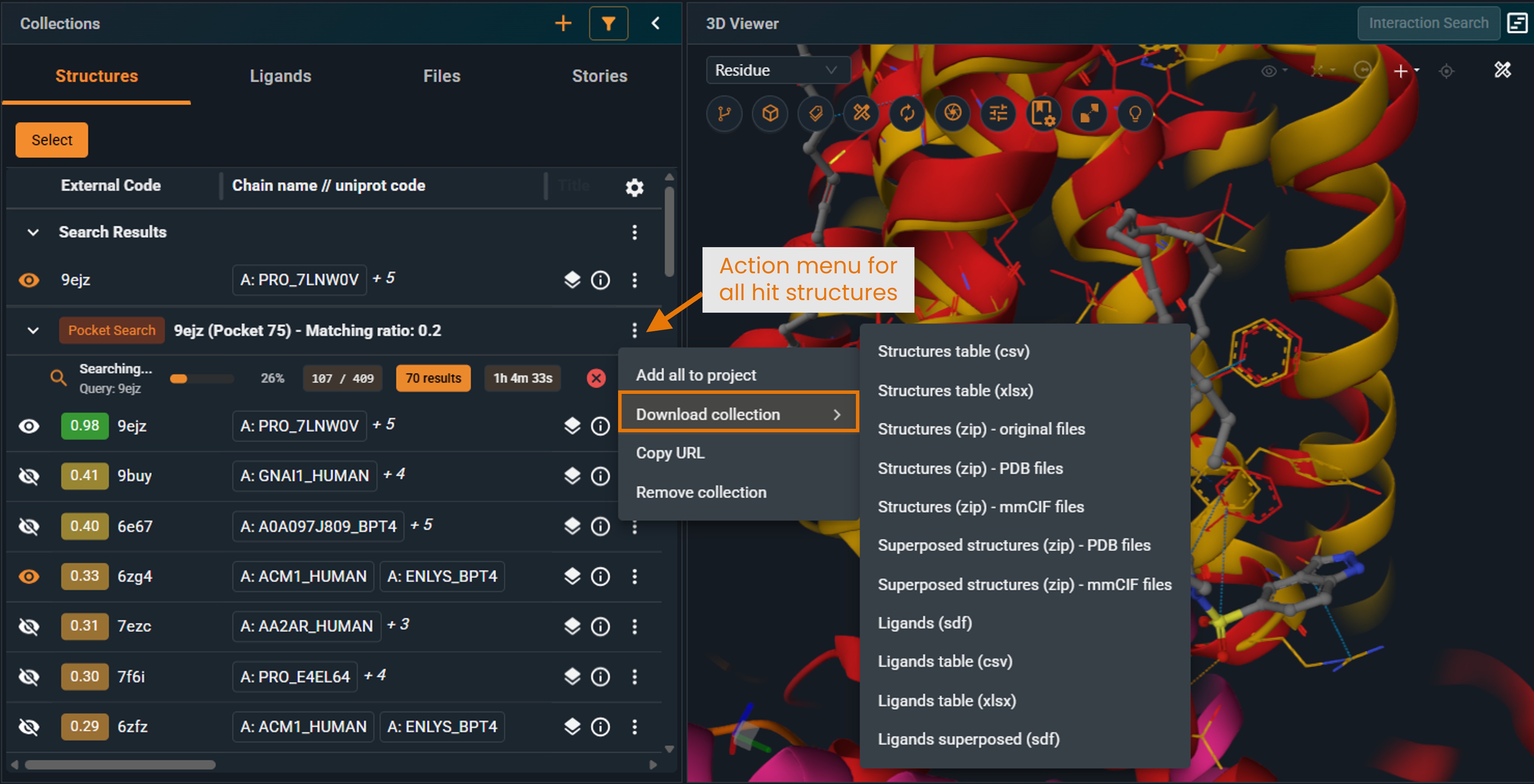 |
|---|
Depending on your search filters, the query structure may not be present in the hit list.
Download a selection of hits
- Click Select (orange button at the top of the Collections panel).
 |
|---|
- Tick the checkboxes for the structures you want to download (make sure to include the query structure for reference).
- In the Selection section, click ACTIONS.
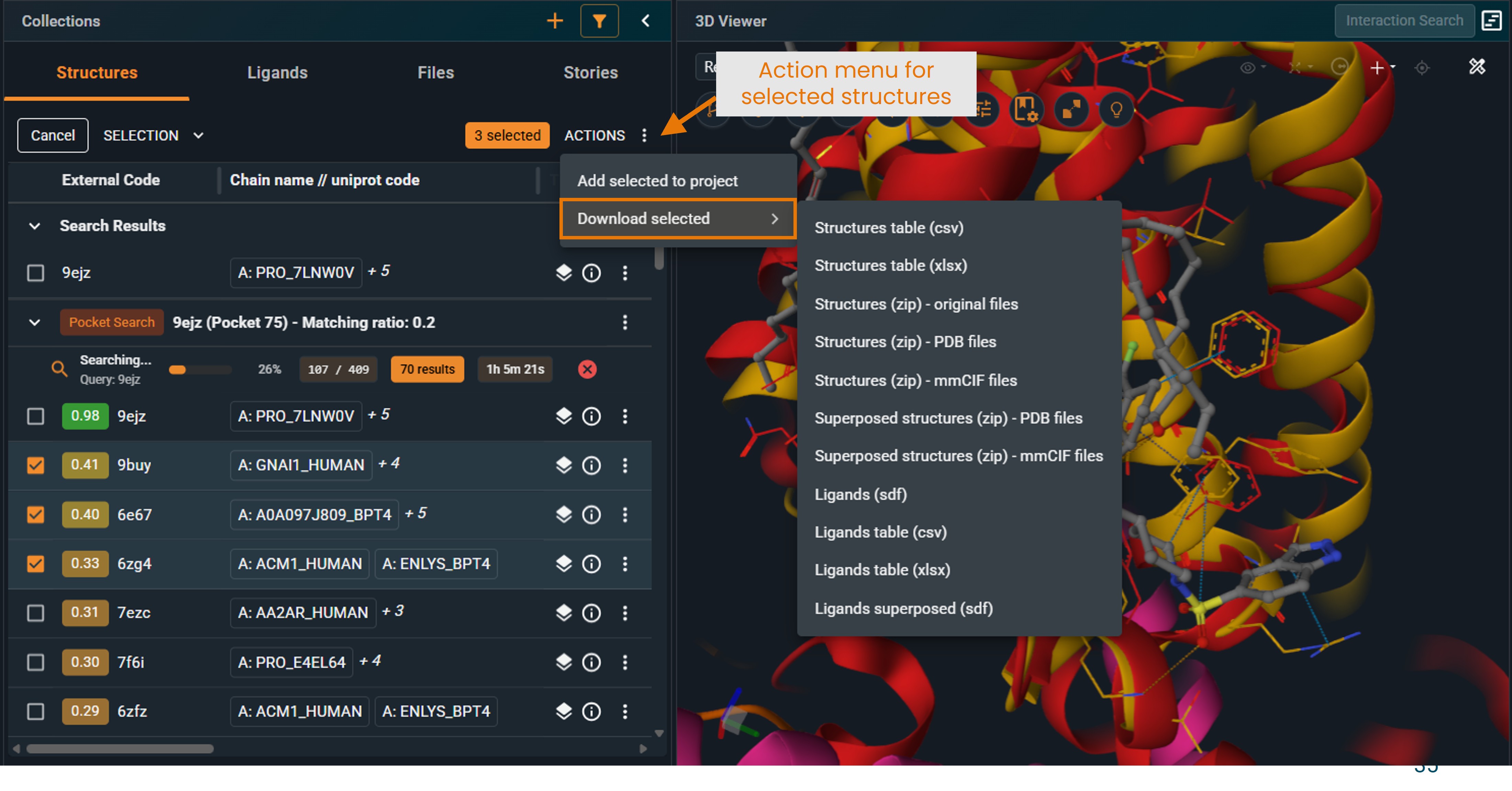 |
|---|
- Select Download selected.
Available download formats
- Structure table (
.csvor.xlsx): List of structure codes retrieved by the search, including properties such as score, structure title, resolution, etc. - Superposed structures (
.zipof.pdbor.mmciffiles): Download the hit structures in the selected format, in the coordinate frame of the superposition.
You do not need to display them in the 3D viewer for them to be downloadable as superposed structures. - Superposed ligands (
.sdf): Ligands found in the hit structures, in the coordinates of the superposed structures.
¶ Legacy UI
Download all results
- Open the action menu of the Pocket Similarity Search result collection.
 |
|---|
- Select Download all X, with X equal to the number of hits of the search.
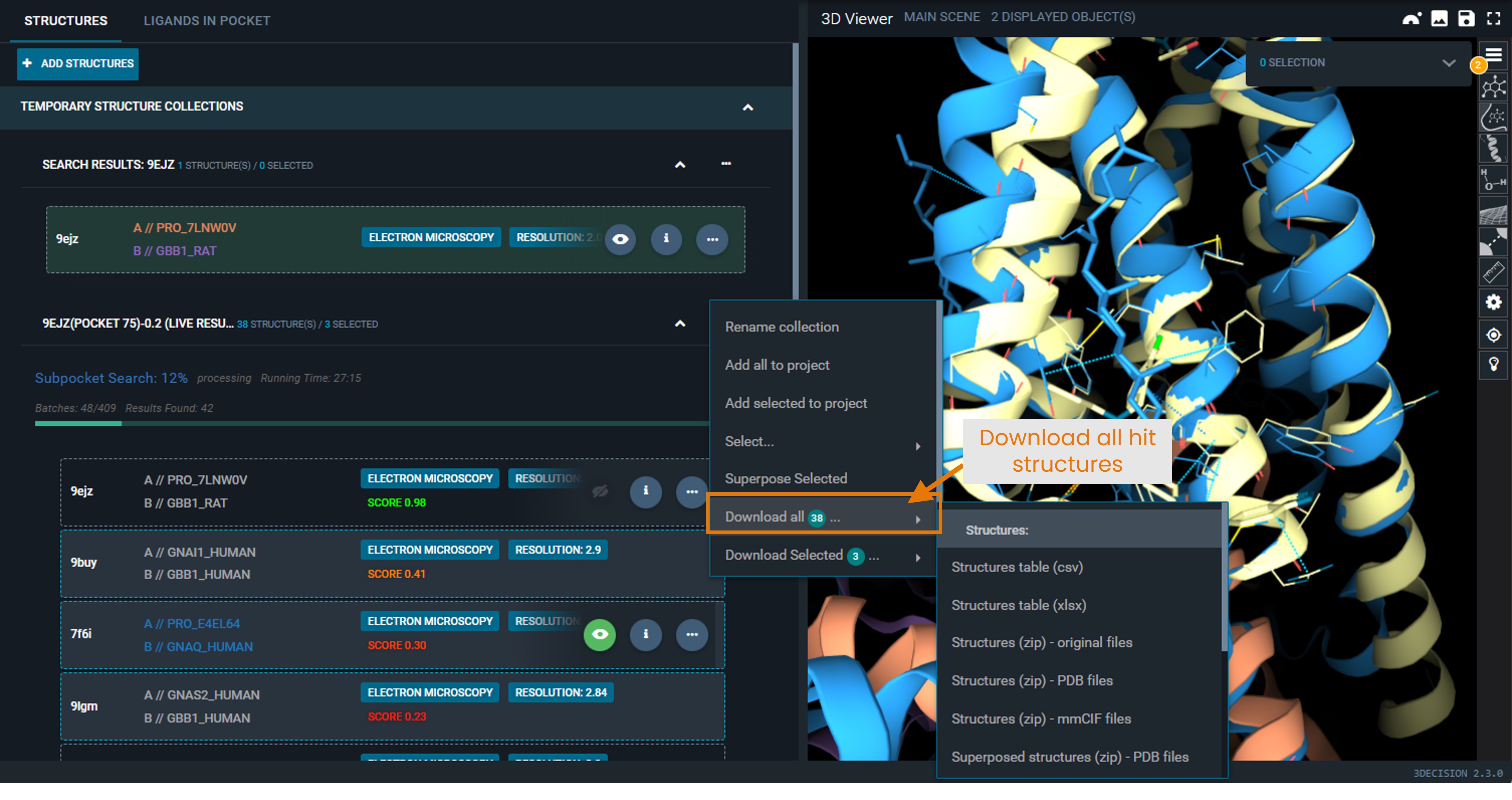 |
|---|
Depending on your search filters, the query structure may not be present in the hit list.
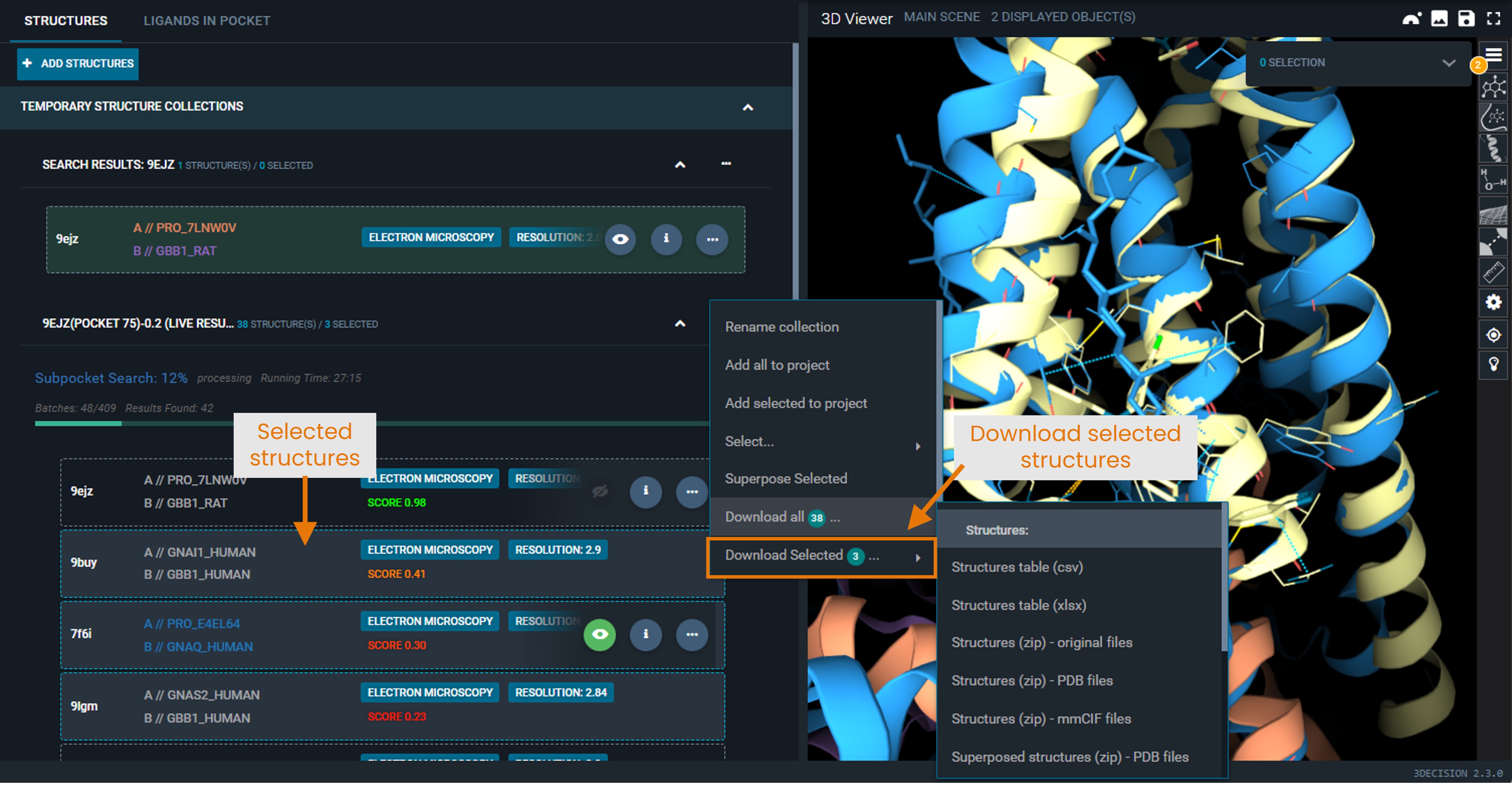
Download a selection of hits
- Single left-click on the structures you want to select. They will be highlighted by a blue border.
- Open the action menu of the Pocket Similarity Search result collection.
- Select Download selected X, with X equal to the number of selected structures .
 |
|---|
Available download formats
- Structure table (
.csvor.xlsx): List of structure codes retrieved by the search, including properties such as score, structure title, resolution, etc. - Superposed structures (
.zipof.pdbor.mmciffiles): Download the hit structures in the selected format, in the coordinate frame of the superposition.
You do not need to display them in the 3D viewer for them to be downloadable as superposed structures. - Superposed ligands (
.sdf): Ligands found in the hit structures, in the coordinates of the superposed structures.
¶ How It Works (Background)
¶ How Binding Sites Are Indexed
3decision automatically processes every structure through three sequential analyses to make binding sites searchable:
¶ 1. Pocket Detection
3decision uses the open-source algorithm fpocket to identify binding sites on the protein surface (POST/pocket-detection/{externalCode} endpoint). Fpocket is a geometry-based pocket detection method built on Voronoi tessellation and alpha spheres.
How fpocket works:
Fpocket relies on alpha spheres—spheres that contact four atoms on their boundary and contain no internal atoms. These alpha spheres represent small vacant spaces on the protein surface. The algorithm then:
- Identifies and filters alpha spheres across the protein surface
- Clusters nearby spheres to identify potential binding pockets
- Scores each pocket based on properties calculated from pocket atoms (such as volume and druggability score)
Fpocket detects pockets both where ligands are present and where they could potentially bind, ensuring comprehensive coverage of the structural database.
¶ 2. Ligand-Cavity Overlap Analysis
For structures containing ligands, 3decision calculates how much each ligand overlaps with detected cavities (POST/ligand-cavity-overlap/{externalCode} endpoint). This analysis enables the "Pockets with ligands only" filter, allowing you to search specifically for binding sites with known chemical matter.
¶ 3. Pocket Feature Analysis
For pockets meeting minimum size criteria (at least 30 alpha spheres or containing any ligand), 3decision characterizes the physicochemical features exposed by lining residues (POST/pocket-features/{externalCode} endpoint).
For each residue in the pocket, the system encodes a 6-bit fingerprint:
- Hydrogen bond acceptor (HBA)
- Hydrogen bond donor (HBD)
- Aromatic (ARO)
- Aliphatic (ALI)
- Positively charged (POS)
- Negatively charged (NEG)
Backbone atoms are not considered when creating the fingerprint of a pocket feature. In the previous Pocket Similarity Search version, backbone atoms were included.
Each fingerprint is stored in the database along with the Cα position of the residue and together they form a pocket feature.
Pocket Feature Pair registration: The system then analyzes pairs of pocket features within each pocket. For pocket features within 10 Å of each other, 3decision computes (1) the distance between the two residues’ Cα atoms and (2) their relative orientation in a local residue frame (an orthonormal basis defined from the backbone atoms N–Cα–C). Together, these geometric descriptors define the vector from one residue’s feature to the other (and vice versa), creating a detailed spatial map of the binding site.
These pocket feature pairs form the searchable signatures used for similarity matching.
¶ How Matching Works
The search identifies similar pockets by comparing the pocket feature pairs in the query pocket with all other indexed pockets in the database (which consists of more than 220k public structures, both experimental from the PDB and AlphaFoldDB human models, plus proprietary structures; each presenting multiple pockets).
The ensemble of pocket feature pairs within a pocket represents both the physicochemical features and their spatial arrangement.
-
Pocket Feature Pair Matching: The algorithm searches for feature pairs whose distances and angles are similar to those in the query pocket. Compared with previous versions, angle comparisons are bidirectional (A→B and B→A). Distance and angle tolerances have been optimized and are now hard-coded (no longer user-configurable).
-
Pocket Matching: A pocket is considered a match if it reproduces a sufficient portion of the query pocket’s feature pairs. In practice, 3decision measures what fraction of the query pocket’s feature pairs can be matched in the candidate pocket, and compares this value to the match score threshold. The default threshold is 0.2, meaning at least 20% of the query feature pairs must have a match.
-
Pocket Feature Clustering: For each matching pocket, 3decision tries to create clusters of matching pocket feature pairs that give a geometrically consistent match (based on RMSD between matching Cα atoms).
-
Similarity Score Calculation: The similarity score is first computed as the ratio between (a) the number of pocket feature pairs in the query pocket that find a match in the hit pocket and (b) the total number of pocket feature pairs in the query pocket. This value is then normalized by the score obtained when comparing the query pocket to itself (query vs. query), which is intended to put scores on a consistent scale (typically between 0 and 1).
In some cases, small deviations can occur because multiple combinations of matching pocket feature pairs can satisfy the matching criteria, and the scoring procedure selects among these possibilities. As a result, it is normal for the query structure compared to itself to yield a score slightly below or above 1 (e.g., 0.98 or 1.01).
Higher scores = more feature pairs match with similar geometry = more similar pockets
¶ Troubleshooting & FAQ
¶ My search is launched but gets stuck at 0% progress
Cause: Your query pocket is probably not indexed correctly.
Solution:
- Open the API Swagger page (bottom-left profile icon → API → 3decision API Endpoint Definition).
- Find the endpoint
POST /pocket-features/{externalCode}. - Click Try it out, enter the structure code, set
committotrue, then click Execute. - Launch the Pocket Similarity Search again.
If it still gets stuck at 0%, contact support.
¶ My search fails immediately
Cause: Your query pocket is probably too small to be indexed, and therefore cannot be used as a search query. Only pockets with more than 30 alpha spheres (roughly a volume above 400 Å) are indexed.
How to verify:
- Open the structure actions menu.
- Click Show pockets.
- Check the Volume value for the pocket.
If the pocket volume looks large enough and the search still fails immediately, contact support.
¶ My search is taking a very long time
This is normal for searches against the full database. The search runs in batches and can take several minutes.
What to do:
- Results appear in real-time—you can start analyzing immediately
- Use the progress bar to monitor completion
- Consider adding filters to reduce search space
- For quick tests, use PDB filter with a small set
¶ Can I save my search results?
Currently, you can:
- Keep the workspace open to preserve your results
- Download structures of interest
Search sessions are not automatically persisted across logins. For API-based workflows, results can be retrieved programmatically.