¶ Introduction
Interaction search is a feature to retrieve structures containing a 3D query pattern of interactions. The matching is based on a selection of interactions in a query system. The user can refine interactants and interaction type, interaction length and orientation tolerance. The returned hits are superimposed to the query and can be displayed in the 3D viewer.
¶ Cautions
This search handles a wide amount of 3D data, and a fuzzy search can run during tens of minutes. A good practice is to start with a precise query as first search, and to make it progressively fuzzier if the number of hits is too limited.
¶ Step-by-step tutorial
This section describes step-by-step how to setup an interaction search and visualize associated results.
¶ Display structure of interest
Steps:
- From the 3decision homepage, search for code 3c79 (crystal structure of acetycholin binding protein with Imidacloprid).
- Click on Open button, on the top right of the search result page.
The structure is loaded and centered on the pocket containing Imidacloprid (IM4). Protein-ligand interactions are displayed in dash lines. All interaction types are not displayed by default to not overcharge the structure view. You can access to the detail of displayed interaction through icon Interactions representation options in the toolbar at the right of the 3D viewer.
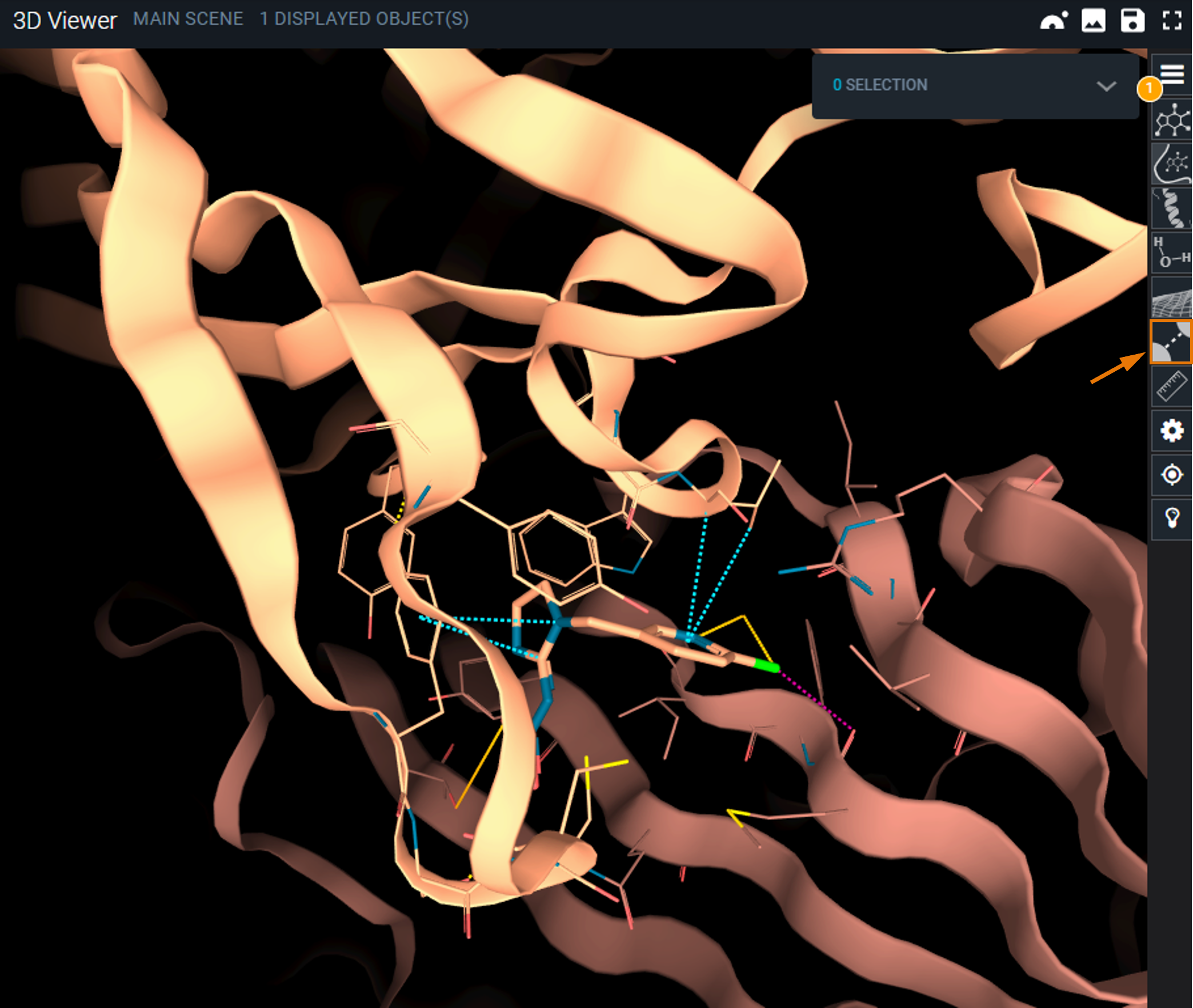
¶ Setup interaction search
For this example, we will setup a simple two interactions search that should be able to gather out-of-the-box scaffolds with a binding mode similar to the one of Imidacloprid.
- Select halogen bond between IM4 chloride and MET116 carbonyl.
- Select H-bond between IM4 NO2 and SER189.
On top right of the 3D Viewer, an expandable panel now mentions 2 selections.
- Click on the selection panel.
- In the expanded panel, click on Interaction Search.
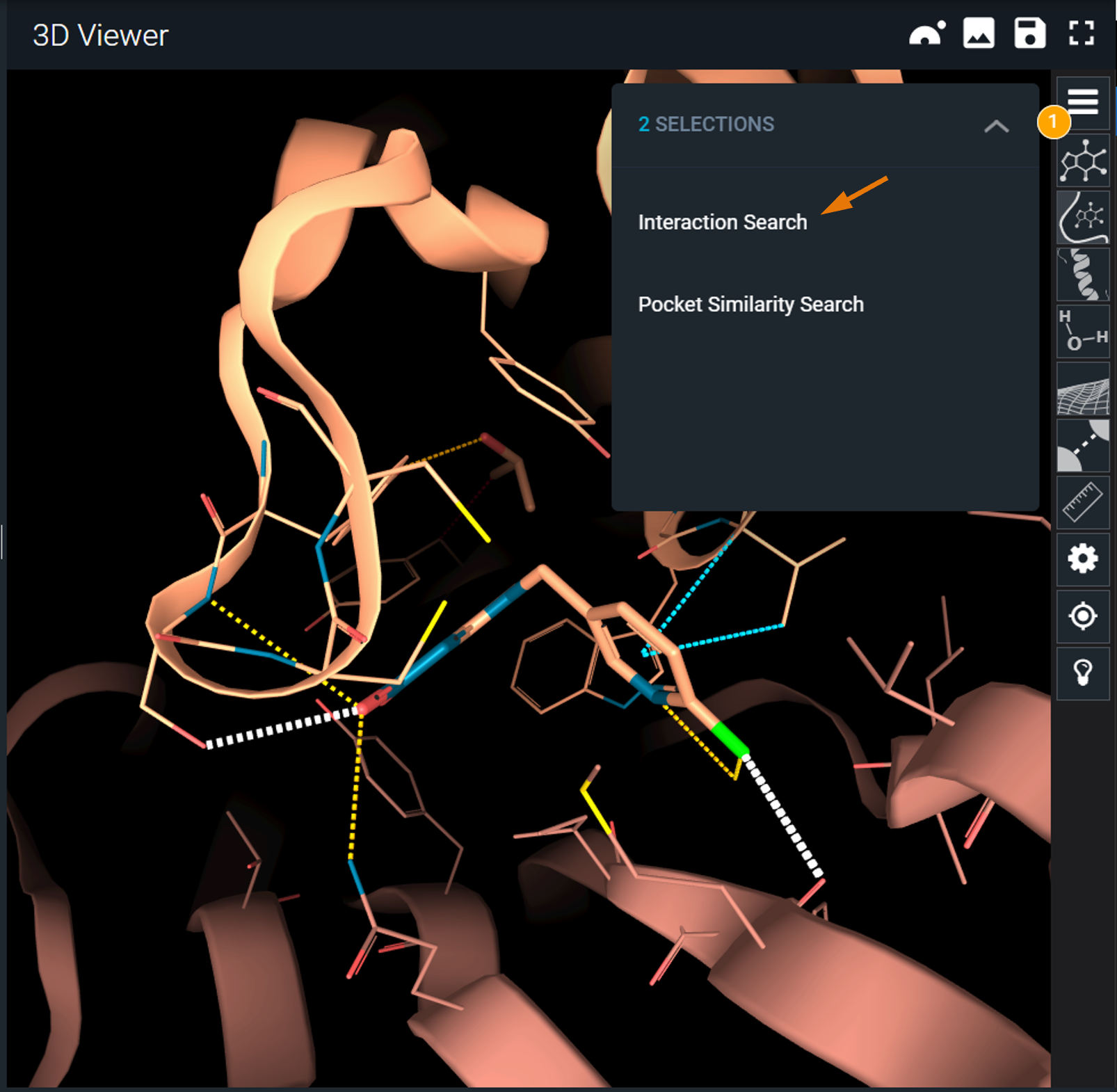
The interaction search panel is now displayed on the left and contains the interactions previously selected.
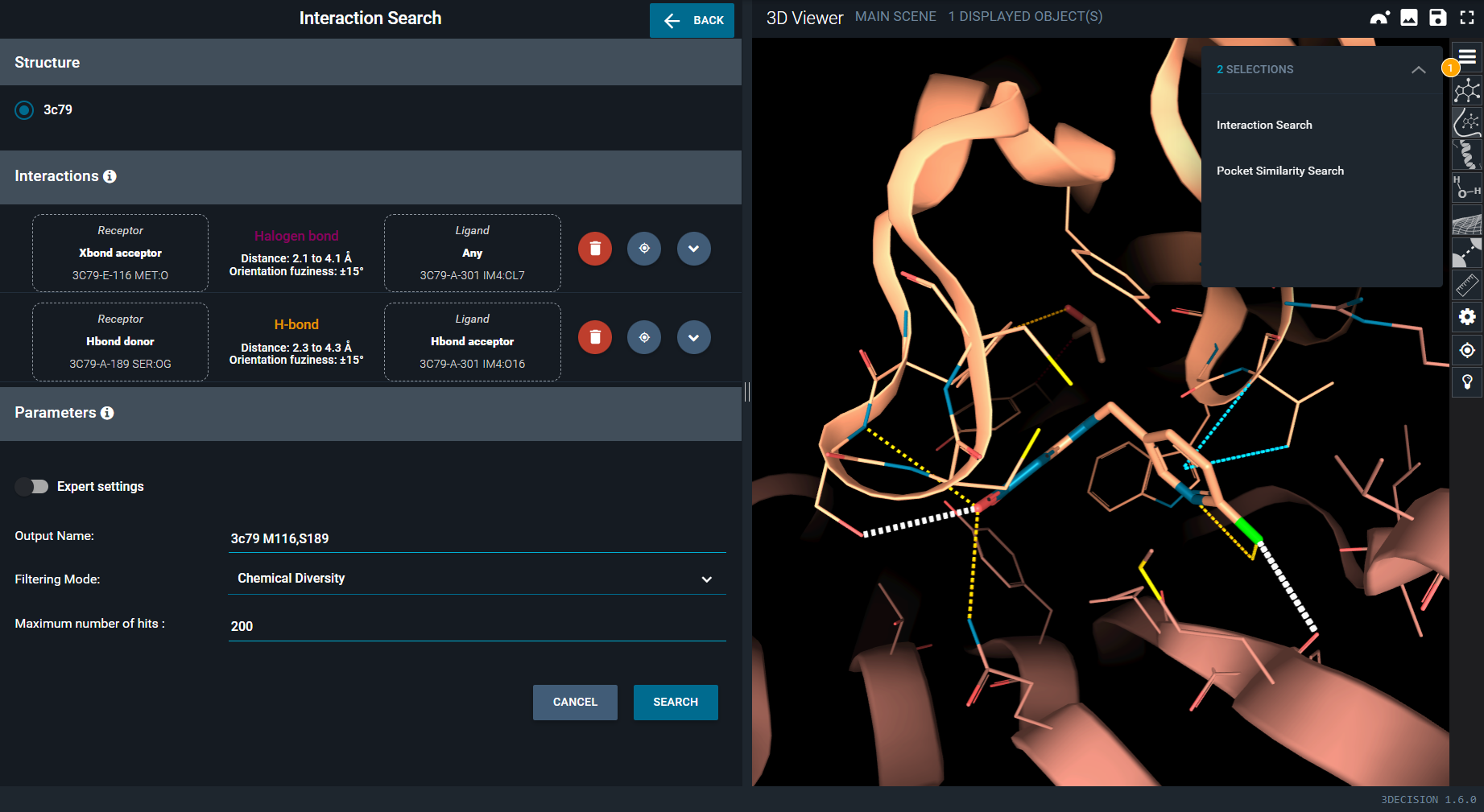
The search can be run with the default parameters, but it is possible to finely tune each interaction parameters. This is what is done in this next steps:
- In the interaction search panel, click on the halogen bond. The subpanel expands and you have acces to the advanced interaction parameters.
- In the drop-down menu under Ligand, select the interactant type xbond donor to ensure an halogen bond donor at the place of IM4 chloride.
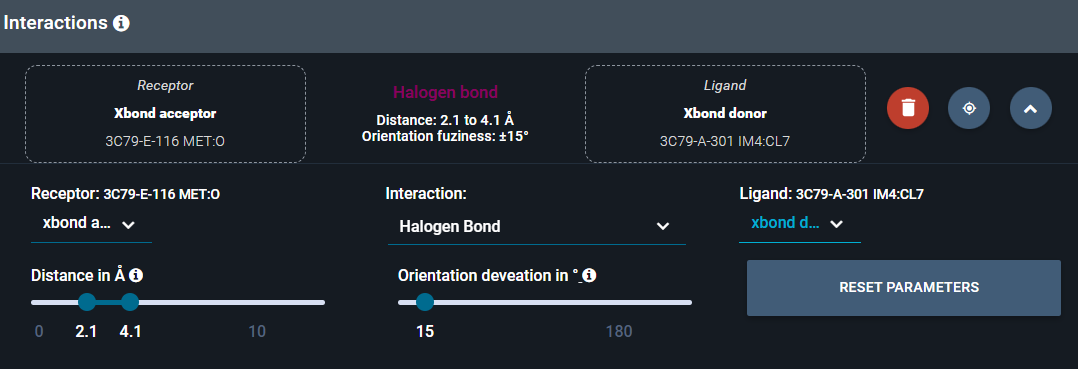
- Click again on the halogen bond in the search panel to close the subpanel.
- Open now the H-bond subpanel and raise the Angle permissivity to 30° to allow a bit more flexibility in H-bond position relatively to the halogen bond.
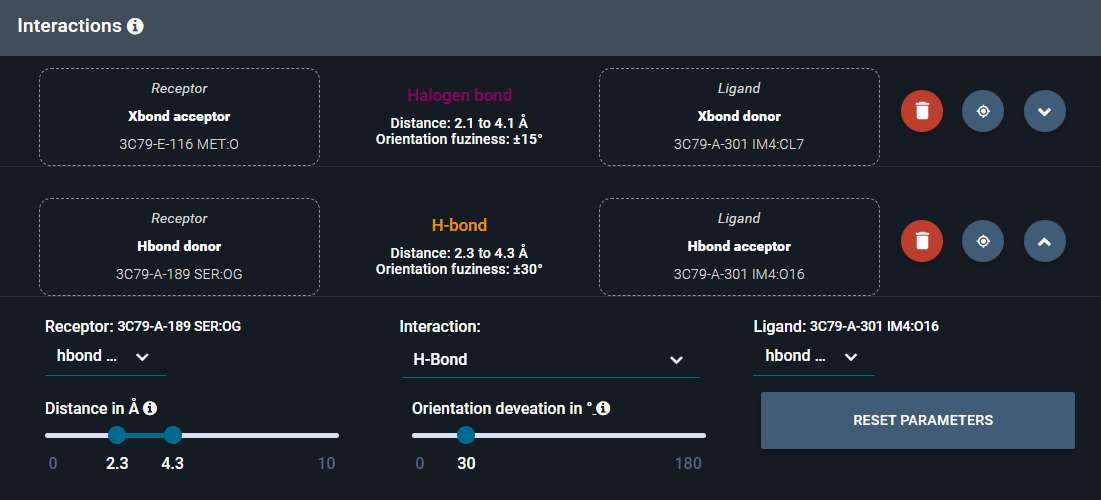
- Close the H-bond subpanel.
- Click on SEARCH blue button, at the bottom of interaction search panel, to run the interaction search.
The search can take a few minutes.
¶ Display a hit
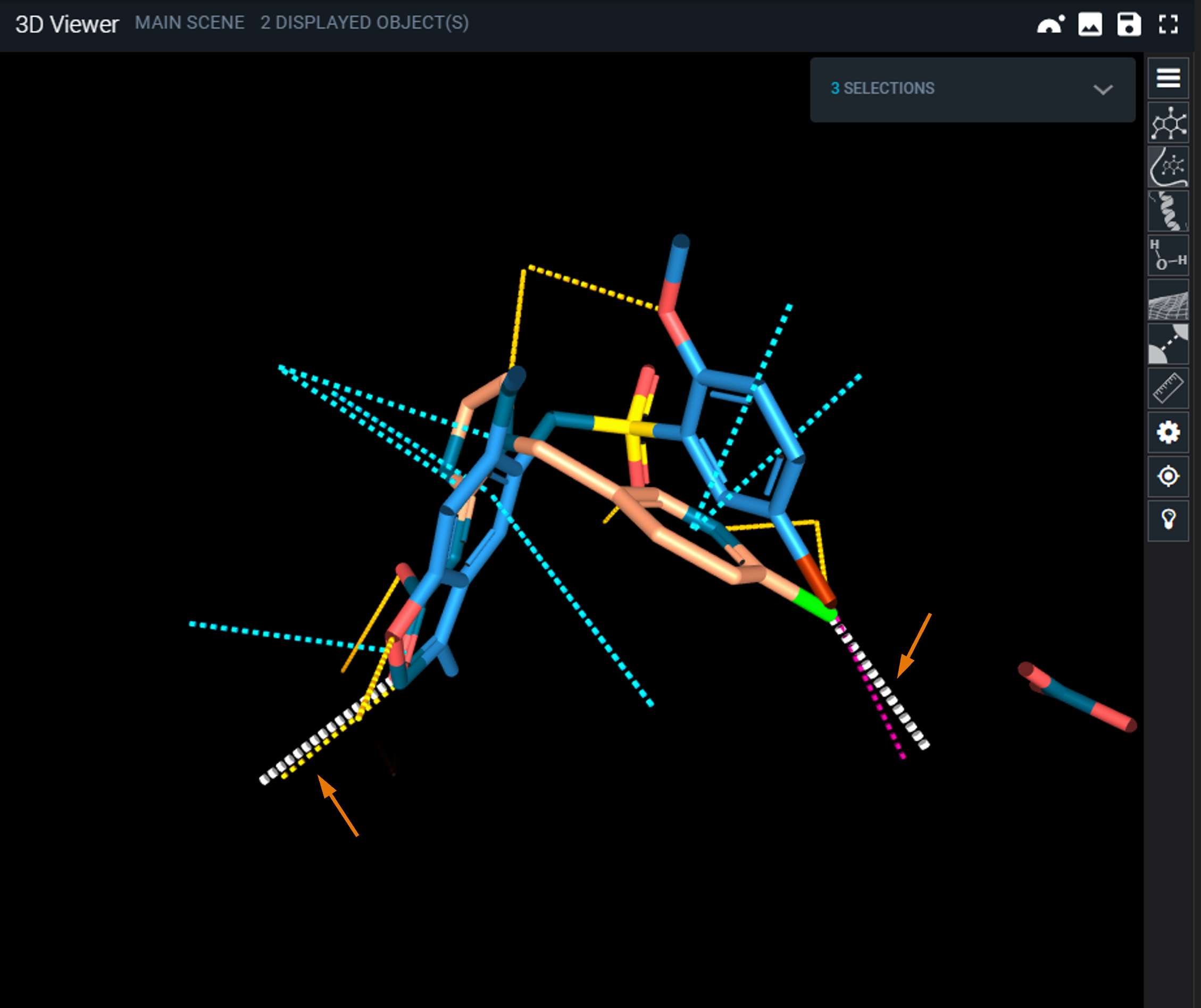
At the end of the search, you are brought to the ligand panel of the workspace, where a new collection containing the interaction search results has been created.
- Hover on the ligand card with the mouse from the collection with the best score and click on the eye icon to display the superimposed hit.
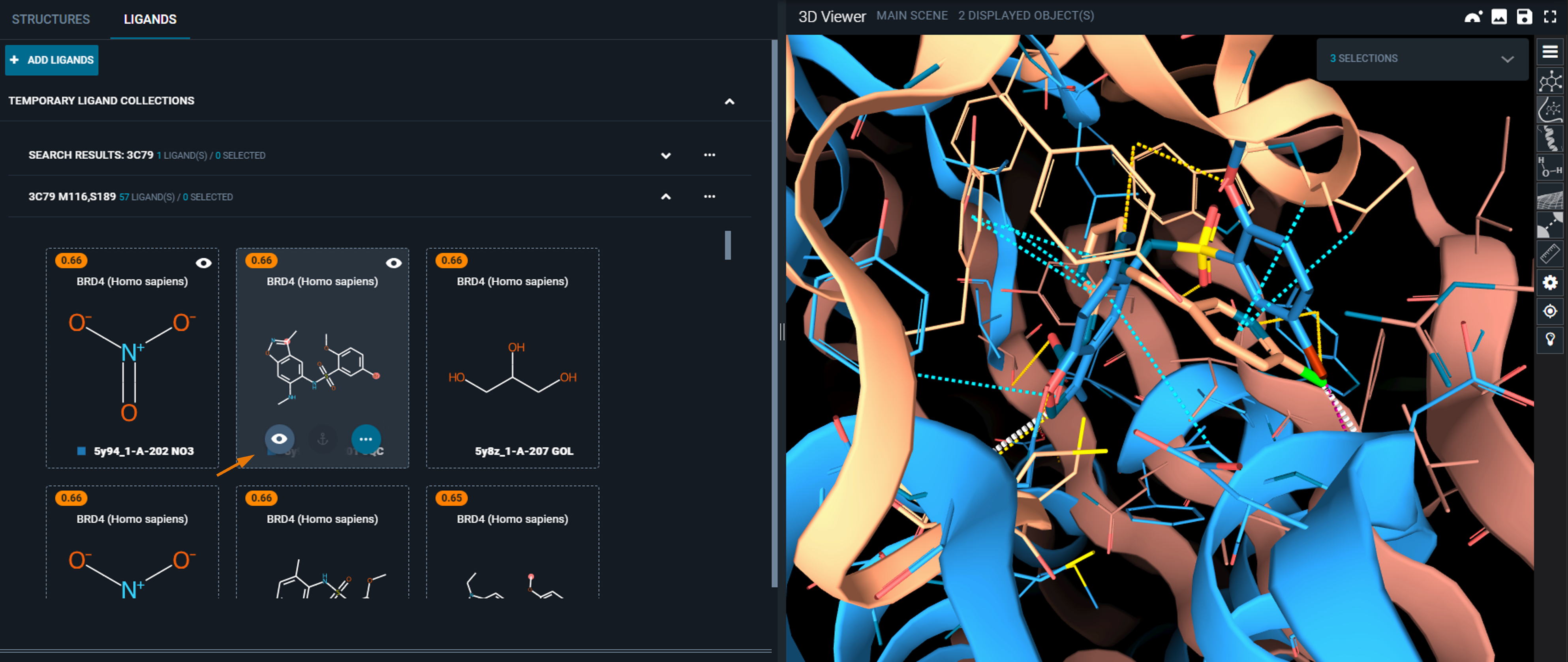
- To clean the view and facilitate the inspection of the binding mode, use the shortcuts on the right toolbar of the 3D Viewer: hide Pockets and Cartoon.
You can now clearly see the query interactions that closely match the interactions in the hit structure.